The mycoplasma surface proteins MIB and MIP promote the dissociation of the antibody-antigen interaction
- PMID: 33674316
- PMCID: PMC7935358
- DOI: 10.1126/sciadv.abf2403
The mycoplasma surface proteins MIB and MIP promote the dissociation of the antibody-antigen interaction
Abstract
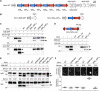
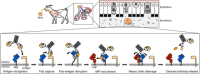
Mycoplasma immunoglobulin binding (MIB) and mycoplasma immunoglobulin protease (MIP) are surface proteins found in the majority of mycoplasma species, acting sequentially to capture antibodies and cleave off their VH domains. Cryo-electron microscopy structures show how MIB and MIP bind to a Fab fragment in a "hug of death" mechanism. As a result, the orientation of the VL and VH domains is twisted out of alignment, disrupting the antigen binding site. We also show that MIB-MIP has the ability to promote the dissociation of the antibody-antigen complex. This system is functional in cells and protects mycoplasmas from antibody-mediated agglutination. These results highlight the key role of the MIB-MIP system in immunity evasion by mycoplasmas through an unprecedented mechanism, and open exciting perspectives to use these proteins as potential tools in the antibody field.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures




References
-
- Kuijl C., Neefjes J., New insight into the everlasting host-pathogen arms race. Nat. Immunol. 10, 808–809 (2009). - PubMed
-
- Finlay B. B., McFadden G., Anti-immunology: Evasion of the host immune system by bacterial and viral pathogens. Cell 124, 767–782 (2006). - PubMed
-
- Sidorin E. V., Solov’eva T. F., IgG-binding proteins of bacteria. Biochemistry (Mosc.) 76, 295–308 (2011). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

