Mitochondrial arginase-2 is essential for IL-10 metabolic reprogramming of inflammatory macrophages
- PMID: 33674584
- PMCID: PMC7936006
- DOI: 10.1038/s41467-021-21617-2
Mitochondrial arginase-2 is essential for IL-10 metabolic reprogramming of inflammatory macrophages
Abstract
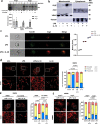
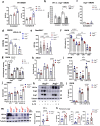
Mitochondria are important regulators of macrophage polarisation. Here, we show that arginase-2 (Arg2) is a microRNA-155 (miR-155) and interleukin-10 (IL-10) regulated protein localized at the mitochondria in inflammatory macrophages, and is critical for IL-10-induced modulation of mitochondrial dynamics and oxidative respiration. Mechanistically, the catalytic activity and presence of Arg2 at the mitochondria is crucial for oxidative phosphorylation. We further show that Arg2 mediates this process by increasing the activity of complex II (succinate dehydrogenase). Moreover, Arg2 is essential for IL-10-mediated downregulation of the inflammatory mediators succinate, hypoxia inducible factor 1α (HIF-1α) and IL-1β in vitro. Accordingly, HIF-1α and IL-1β are highly expressed in an LPS-induced in vivo model of acute inflammation using Arg2-/- mice. These findings shed light on a new arm of IL-10-mediated metabolic regulation, working to resolve the inflammatory status of the cell.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

