lncRNA transcription induces meiotic recombination through chromatin remodelling in fission yeast
- PMID: 33674718
- PMCID: PMC7935937
- DOI: 10.1038/s42003-021-01798-8
lncRNA transcription induces meiotic recombination through chromatin remodelling in fission yeast
Abstract
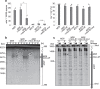
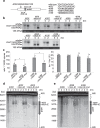
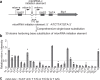
Noncoding RNAs (ncRNAs) are involved in various biological processes, including gene expression, development, and disease. Here, we identify a novel consensus sequence of a cis-element involved in long ncRNA (lncRNA) transcription and demonstrate that lncRNA transcription from this cis-element activates meiotic recombination via chromatin remodeling. In the fission yeast fbp1 gene, glucose starvation induces a series of promoter-associated lncRNAs, referred to as metabolic-stress-induced lncRNAs (mlonRNAs), which contribute to chromatin remodeling and fbp1 activation. Translocation of the cis-element required for mlonRNA into a well-characterized meiotic recombination hotspot, ade6-M26, further stimulates transcription and meiotic recombination via local chromatin remodeling. The consensus sequence of this cis-element (mlon-box) overlaps with meiotic recombination sites in the fission yeast genome. At one such site, the SPBC24C6.09c upstream region, meiotic double-strand break (DSB) formation is induced in an mlon-box-dependent manner. Therefore, mlonRNA transcription plays a universal role in chromatin remodeling and the regulation of transcription and recombination.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Multi-Layered Regulations on the Chromatin Architectures: Establishing the Tight and Specific Responses of Fission Yeast fbp1 Gene Transcription.Biomolecules. 2022 Nov 5;12(11):1642. doi: 10.3390/biom12111642. Biomolecules. 2022. PMID: 36358992 Free PMC article. Review.
-
lncRNA transcriptional initiation induces chromatin remodeling within a limited range in the fission yeast fbp1 promoter.Sci Rep. 2019 Jan 22;9(1):299. doi: 10.1038/s41598-018-36049-0. Sci Rep. 2019. PMID: 30670704 Free PMC article.
-
Fission yeast Tup1-like repressors repress chromatin remodeling at the fbp1+ promoter and the ade6-M26 recombination hotspot.Genetics. 2003 Oct;165(2):505-15. doi: 10.1093/genetics/165.2.505. Genetics. 2003. PMID: 14573465 Free PMC article.
-
Multiple modes of chromatin configuration at natural meiotic recombination hot spots in fission yeast.Eukaryot Cell. 2007 Nov;6(11):2072-80. doi: 10.1128/EC.00246-07. Epub 2007 Sep 7. Eukaryot Cell. 2007. PMID: 17827346 Free PMC article.
-
Regulation Mechanisms of Meiotic Recombination Revealed from the Analysis of a Fission Yeast Recombination Hotspot ade6-M26.Biomolecules. 2022 Nov 26;12(12):1761. doi: 10.3390/biom12121761. Biomolecules. 2022. PMID: 36551189 Free PMC article. Review.
Cited by
-
Construction of a human hTERT RPE-1 cell line with inducible Cre for editing of endogenous genes.Biol Open. 2022 Feb 15;11(2):bio059056. doi: 10.1242/bio.059056. Epub 2022 Feb 16. Biol Open. 2022. PMID: 35067715 Free PMC article.
-
Reciprocal stabilization of transcription factor binding integrates two signaling pathways to regulate fission yeast fbp1 transcription.Nucleic Acids Res. 2021 Sep 27;49(17):9809-9820. doi: 10.1093/nar/gkab758. Nucleic Acids Res. 2021. PMID: 34486060 Free PMC article.
-
Metabolic stress-induced long ncRNA transcription governs the formation of meiotic DNA breaks in the fission yeast fbp1 gene.PLoS One. 2024 Jan 22;19(1):e0294191. doi: 10.1371/journal.pone.0294191. eCollection 2024. PLoS One. 2024. PMID: 38252660 Free PMC article.
-
Multi-Layered Regulations on the Chromatin Architectures: Establishing the Tight and Specific Responses of Fission Yeast fbp1 Gene Transcription.Biomolecules. 2022 Nov 5;12(11):1642. doi: 10.3390/biom12111642. Biomolecules. 2022. PMID: 36358992 Free PMC article. Review.
-
Identification and functional analysis of growth rate associated long non-coding RNAs in Komagataella phaffii.Comput Struct Biotechnol J. 2025 Apr 22;27:1693-1705. doi: 10.1016/j.csbj.2025.04.028. eCollection 2025. Comput Struct Biotechnol J. 2025. PMID: 40352477 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

