IFN signaling and neutrophil degranulation transcriptional signatures are induced during SARS-CoV-2 infection
- PMID: 33674719
- PMCID: PMC7935909
- DOI: 10.1038/s42003-021-01829-4
IFN signaling and neutrophil degranulation transcriptional signatures are induced during SARS-CoV-2 infection
Abstract
SARS-CoV-2 virus has infected more than 92 million people worldwide resulting in the Coronavirus disease 2019 (COVID-19). Using a rhesus macaque model of SARS-CoV-2 infection, we have characterized the transcriptional signatures induced in the lungs of juvenile and old macaques following infection. Genes associated with Interferon (IFN) signaling, neutrophil degranulation and innate immune pathways are significantly induced in macaque infected lungs, while pathways associated with collagen formation are downregulated, as also seen in lungs of macaques with tuberculosis. In COVID-19, increasing age is a significant risk factor for poor prognosis and increased mortality. Type I IFN and Notch signaling pathways are significantly upregulated in lungs of juvenile infected macaques when compared with old infected macaques. These results are corroborated with increased peripheral neutrophil counts and neutrophil lymphocyte ratio in older individuals with COVID-19 disease. Together, our transcriptomic studies have delineated disease pathways that improve our understanding of the immunopathogenesis of COVID-19.
Conflict of interest statement
The authors declare no competing interests.
Figures
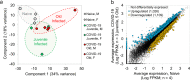



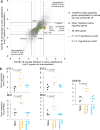





Update of
-
IFN signaling and neutrophil degranulation transcriptional signatures are induced during SARS-CoV-2 infection.bioRxiv [Preprint]. 2020 Aug 6:2020.08.06.239798. doi: 10.1101/2020.08.06.239798. bioRxiv. 2020. Update in: Commun Biol. 2021 Mar 5;4(1):290. doi: 10.1038/s42003-021-01829-4. PMID: 32793903 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

