Kinetic modeling of anaerobic degradation of plant-derived aromatic mixtures by Rhodopseudomonas palustris
- PMID: 33675449
- PMCID: PMC7997838
- DOI: 10.1007/s10532-021-09932-3
Kinetic modeling of anaerobic degradation of plant-derived aromatic mixtures by Rhodopseudomonas palustris
Abstract
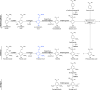
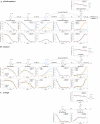
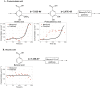
Rhodopseudomonas palustris is a model microorganism for studying the anaerobic metabolism of aromatic compounds. While it is well documented which aromatics can serve as sole organic carbon sources, co-metabolism of other aromatics is poorly understood. This study used kinetic modeling to analyze the simultaneous degradation of aromatic compounds present in corn stover hydrolysates and model the co-metabolism of aromatics not known to support growth of R. palustris as sole organic substrates. The simulation predicted that p-coumaroyl amide and feruloyl amide were hydrolyzed to p-coumaric acid and ferulic acid, respectively, and further transformed via p-coumaroyl-CoA and feruloyl-CoA. The modeling also suggested that metabolism of p-hydroxyphenyl aromatics was slowed by substrate inhibition, whereas the transformation of guaiacyl aromatics was inhibited by their p-hydroxyphenyl counterparts. It also predicted that substrate channeling may occur during degradation of p-coumaroyl-CoA and feruloyl-CoA, resulting in no detectable accumulation of p-hydroxybenzaldehyde and vanillin, during the transformation of these CoA ligated compounds to p-hydroxybenzoic acid and vanillic acid, respectively. While the simulation correctly represented the known transformation of p-hydroxybenzoic acid via the benzoyl-CoA pathway, it also suggested co-metabolism of vanillic acid and syringic acid, which are known not to serve as photoheterotrophic growth substrate for R. palustris.
Keywords: Anaerobic degradation; Co-metabolism; Kinetic modeling; Plant-derived aromatics; Rhodopseudomonas palustris; Substrate inhibition.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Similar articles
-
Anaerobic Degradation of Syringic Acid by an Adapted Strain of Rhodopseudomonas palustris.Appl Environ Microbiol. 2020 Jan 21;86(3):e01888-19. doi: 10.1128/AEM.01888-19. Print 2020 Jan 21. Appl Environ Microbiol. 2020. PMID: 31732577 Free PMC article.
-
Metabolism of Multiple Aromatic Compounds in Corn Stover Hydrolysate by Rhodopseudomonas palustris.Environ Sci Technol. 2015 Jul 21;49(14):8914-22. doi: 10.1021/acs.est.5b02062. Epub 2015 Jul 9. Environ Sci Technol. 2015. PMID: 26121369 Free PMC article.
-
Benzoyl coenzyme a pathway-mediated metabolism of meta-hydroxy-aromatic acids in Rhodopseudomonas palustris.J Bacteriol. 2013 Sep;195(18):4112-20. doi: 10.1128/JB.00634-13. Epub 2013 Jul 12. J Bacteriol. 2013. PMID: 23852864 Free PMC article.
-
The bacterial degradation of benzoic acid and benzenoid compounds under anaerobic conditions: unifying trends and new perspectives.FEMS Microbiol Rev. 1994 Apr;13(4):441-68. doi: 10.1111/j.1574-6976.1994.tb00061.x. FEMS Microbiol Rev. 1994. PMID: 8011356 Review.
-
Anaerobic biodegradation of aromatic compounds.Indian J Exp Biol. 2003 Sep;41(9):1046-67. Indian J Exp Biol. 2003. PMID: 15242297 Review.
Cited by
-
Thioredoxin Reductase-Type Ferredoxin: NADP+ Oxidoreductase of Rhodopseudomonas palustris: Potentiometric Characteristics and Reactions with Nonphysiological Oxidants.Antioxidants (Basel). 2022 May 19;11(5):1000. doi: 10.3390/antiox11051000. Antioxidants (Basel). 2022. PMID: 35624864 Free PMC article.
-
The genome-scale metabolic model for the purple non-sulfur bacterium Rhodopseudomonas palustris Bis A53 accurately predicts phenotypes under chemoheterotrophic, chemoautotrophic, photoheterotrophic, and photoautotrophic growth conditions.PLoS Comput Biol. 2023 Aug 9;19(8):e1011371. doi: 10.1371/journal.pcbi.1011371. eCollection 2023 Aug. PLoS Comput Biol. 2023. PMID: 37556472 Free PMC article.
-
High enzyme promiscuity in lignin degradation mechanisms in Rhodopseudomonas palustris CGA009.Appl Environ Microbiol. 2025 Aug 20;91(8):e0057325. doi: 10.1128/aem.00573-25. Epub 2025 Jul 8. Appl Environ Microbiol. 2025. PMID: 40626778 Free PMC article.
References
-
- Atkinson KA. An Introduction to numerical analysis. 2. New York: John Wiley and Sons; 1989.
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

