Carbon catabolite regulation of secondary metabolite formation, an old but not well-established regulatory system
- PMID: 33675560
- PMCID: PMC8966007
- DOI: 10.1111/1751-7915.13791
Carbon catabolite regulation of secondary metabolite formation, an old but not well-established regulatory system
Abstract
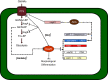
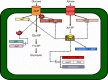
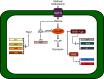
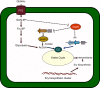
Secondary microbial metabolites have various functions for the producer microorganisms, which allow them to interact and survive in adverse environments. In addition to these functions, other biological activities may have clinical relevance, as diverse as antimicrobial, anticancer and hypocholesterolaemic effects. These metabolites are usually formed during the idiophase of growth and have a wide diversity in their chemical structures. Their synthesis is under the impact of the type and concentration of the culture media nutrients. Some of the molecular mechanisms that affect the synthesis of secondary metabolites in bacteria (Gram-positive and negative) and fungi are partially known. Moreover, all microorganisms have their peculiarities in the control mechanisms of carbon sources, even those belonging to the same genus. This regulatory knowledge is necessary to establish culture conditions and manipulation methods for genetic improvement and product fermentation. As the carbon source is one of the essential nutritional factors for antibiotic production, its study has been imperative both at the industrial and research levels. This review aims to draw the utmost recent advances performed to clarify the molecular mechanisms of the negative effect exerted by the carbon source on the secondary metabolite formation, emphasizing those found in Streptomyces, one of the genera most profitable antibiotic producers.
© 2021 The Authors. Microbial Biotechnology published by John Wiley & Sons Ltd and Society for Applied Microbiology.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures
Similar articles
-
Production of microbial secondary metabolites: regulation by the carbon source.Crit Rev Microbiol. 2010 May;36(2):146-67. doi: 10.3109/10408410903489576. Crit Rev Microbiol. 2010. PMID: 20210692 Review.
-
Interplay between carbon, nitrogen and phosphate utilization in the control of secondary metabolite production in Streptomyces.Antonie Van Leeuwenhoek. 2018 May;111(5):761-781. doi: 10.1007/s10482-018-1073-1. Epub 2018 Mar 31. Antonie Van Leeuwenhoek. 2018. PMID: 29605896 Review.
-
Carbon catabolite regulation in Streptomyces: new insights and lessons learned.World J Microbiol Biotechnol. 2017 Sep;33(9):162. doi: 10.1007/s11274-017-2328-0. Epub 2017 Aug 2. World J Microbiol Biotechnol. 2017. PMID: 28770367 Review.
-
Carbon source regulation of antibiotic production.J Antibiot (Tokyo). 2010 Aug;63(8):442-59. doi: 10.1038/ja.2010.78. Epub 2010 Jul 28. J Antibiot (Tokyo). 2010. PMID: 20664603 Review.
-
Tailored culture strategies to promote antimicrobial secondary metabolite production in Diaporthe caliensis: a metabolomic approach.Microb Cell Fact. 2024 Dec 5;23(1):328. doi: 10.1186/s12934-024-02567-y. Microb Cell Fact. 2024. PMID: 39639292 Free PMC article.
Cited by
-
Dissecting the role of the two Streptomyces peucetius var. caesius glucokinases in the sensitivity to carbon catabolite repression.J Ind Microbiol Biotechnol. 2021 Dec 23;48(9-10):kuab047. doi: 10.1093/jimb/kuab047. J Ind Microbiol Biotechnol. 2021. PMID: 34383077 Free PMC article.
-
Coupled strategy based on regulator manipulation and medium optimization empowers the biosynthetic overproduction of lincomycin.Synth Syst Biotechnol. 2024 Jan 14;9(1):134-143. doi: 10.1016/j.synbio.2024.01.004. eCollection 2024 Mar. Synth Syst Biotechnol. 2024. PMID: 38318491 Free PMC article.
-
Optimized carotenoid production and antioxidant capacity of Gordonia hongkongensis.Sci Prog. 2024 Apr-Jun;107(2):368504241253695. doi: 10.1177/00368504241253695. Sci Prog. 2024. PMID: 38801654 Free PMC article.
-
Effects of Carbon, Nitrogen, Ambient pH and Light on Mycelial Growth, Sporulation, Sorbicillinoid Biosynthesis and Related Gene Expression in Ustilaginoidea virens.J Fungi (Basel). 2023 Mar 23;9(4):390. doi: 10.3390/jof9040390. J Fungi (Basel). 2023. PMID: 37108845 Free PMC article.
-
Differential anaerobic oxidation of benzoate in Geotalea daltonii FRC-32.Microbiol Spectr. 2025 Apr;13(4):e0232424. doi: 10.1128/spectrum.02324-24. Epub 2025 Mar 5. Microbiol Spectr. 2025. PMID: 40042335 Free PMC article.
References
-
- Angell, S. , Lewis, C.G. , Buttner, M.J. , and Bibb, M.J. (1994) Glucose repression in Streptomyces coelicolor A3(2): a likely regulatory role for glucose kinase. Mol Gen Genet 244: 135–143. - PubMed
-
- Angell, S. , Schwarz, E. , and Bibb, M.J. (1992) The glucose kinase gene of Streptomyces coelicolor A3(2): its nucleotide sequence, transcriptional analysis and role in glucose repression. Mol Microbiol 6: 2833–2844. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

