Whole genome de novo sequencing and comparative genomic analyses suggests that Chlamydia psittaci strain 84/2334 should be reclassified as Chlamydia abortus species
- PMID: 33676404
- PMCID: PMC7937271
- DOI: 10.1186/s12864-021-07477-6
Whole genome de novo sequencing and comparative genomic analyses suggests that Chlamydia psittaci strain 84/2334 should be reclassified as Chlamydia abortus species
Abstract
Background: Chlamydia abortus and Chlamydia psittaci are important pathogens of livestock and avian species, respectively. While C. abortus is recognized as descended from C. psittaci species, there is emerging evidence of strains that are intermediary between the two species, suggesting they are recent evolutionary ancestors of C. abortus. Such strains include C. psittaci strain 84/2334 that was isolated from a parrot. Our aim was to classify this strain by sequencing its genome and explore its evolutionary relationship to both C. abortus and C. psittaci.
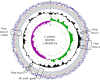
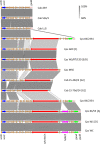
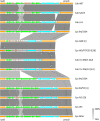
Results: In this study, methods based on multi-locus sequence typing (MLST) of seven housekeeping genes and on typing of five species discriminant proteins showed that strain 84/2334 clustered with C. abortus species. Furthermore, whole genome de novo sequencing of the strain revealed greater similarity to C. abortus in terms of GC content, while 16S rRNA and whole genome phylogenetic analysis, as well as network and recombination analysis showed that the strain clusters more closely with C. abortus strains. The analysis also suggested a closer evolutionary relationship between this strain and the major C. abortus clade, than to two other intermediary avian C. abortus strains or C. psittaci strains. Molecular analyses of genes (polymorphic membrane protein and transmembrane head protein genes) and loci (plasticity zone), found in key virulence-associated regions that exhibit greatest diversity within and between chlamydial species, reveal greater diversity than present in sequenced C. abortus genomes as well as similar features to both C. abortus and C. psittaci species. The strain also possesses an extrachromosomal plasmid, as found in most C. psittaci species but absent from all sequenced classical C. abortus strains.
Conclusion: Overall, the results show that C. psittaci strain 84/2334 clusters very closely with C. abortus strains, and are consistent with the strain being a recent C. abortus ancestral species. This suggests that the strain should be reclassified as C. abortus. Furthermore, the identification of a C. abortus strain bearing an extra-chromosomal plasmid has implications for plasmid-based transformation studies to investigate gene function as well as providing a potential route for the development of a next generation vaccine to protect livestock from C. abortus infection.
Keywords: Chlamydia abortus; Chlamydia psittaci; Comparative genomic analysis; Genome sequence; MLST; Phylogenomics; Plasticity zone; Polymorphic membrane proteins.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Everett KDE, Bush RM, Andersen AA. Emended description of the order Chlamydiales, proposal of ParaChlamydiaceae fam. Nov. and Simkaniaceae fam. Nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int J Syst Bacteriol. 1999;49(Pt2):415–440. doi: 10.1099/00207713-49-2-415. - DOI - PubMed
-
- Horn M. Phylum XXIV. Chlamydiae Garrity and Holt 2001. In: Krieg NR, Staley JT, Brown DR, Hedlund BP, Paster BJ, Ward NL, Ludwig W, Whitman WB, editors. Bergey's manual of systematic bacteriology. 4. 2. New York: Springer; 2011. pp. 843–878.
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

