Effect of fibronectin, FGF-2, and BMP4 in the stemness maintenance of BMSCs and the metabolic and proteomic cues involved
- PMID: 33676544
- PMCID: PMC7936451
- DOI: 10.1186/s13287-021-02227-7
Effect of fibronectin, FGF-2, and BMP4 in the stemness maintenance of BMSCs and the metabolic and proteomic cues involved
Abstract
Background: Growing evidence suggests that the pluripotent state of mesenchymal stem cells (MSCs) relies on specific local microenvironmental cues such as adhesion molecules and growth factors. Fibronectin (FN), fibroblast growth factor 2 (FGF2), and bone morphogenetic protein 4 (BMP4) are the key players in the regulation of stemness and lineage commitment of MSCs. Therefore, this study was designed to investigate the pluripotency and multilineage differentiation of bone marrow-derived MSCs (BMSCs) with the introduction of FN, FGF-2, and BMP4 and to identify the metabolic and proteomic cues involved in stemness maintenance.
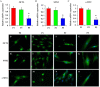
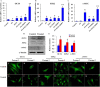
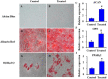
Methods: To elucidate the stemness of BMSCs when treated with FN, FGF-2, and BMP4, the pluripotency markers of OCT4, SOX2, and c-MYC in BMSCs were monitored by real-time PCR and/or western blot. The nuclear translocation of OCT4, SOX2, and c-MYC was investigated by immunofluorescence staining. Multilineage differentiation of the treated BMSCs was determined by relevant differentiation markers. To identify the molecular signatures of BMSC stemness, gas chromatography-mass spectrometry (GC-MS), liquid chromatography-tandem mass spectrometry (LC-MS/MS), and bioinformatics analysis were utilized to determine the metabolite and protein profiles associated with stem cell maintenance.
Results: Our results demonstrated that the expression of stemness markers decreased with BMSC passaging, and the manipulation of the microenvironment with fibronectin and growth factors (FGF2 and BMP4) can significantly improve BMSC stemness. Of note, we revealed 7 differentially expressed metabolites, the target genes of these metabolites may have important implications in the maintenance of BMSCs through their effects on metabolic activity, energy production, and potentially protein production. We also identified 21 differentially abundant proteins, which involved in multiple pathways, including metabolic, autophagy-related, and signaling pathways regulating the pluripotency of stem cells. Additionally, bioinformatics analysis comfirned the correlation between metabolic and proteomic profiling, suggesting that the importance of metabolism and proteome networks and their reciprocal communication in the preservation of stemness.
Conclusions: These results indicate that the culture environment supplemented with the culture cocktail (FN, FGF2, and BMP4) plays an essential role in shaping the pluripotent state of BMSCs. Both the metabolism and proteome networks are involved in this process and the modulation of cell-fate decision making. All these findings may contribute to the application of MSCs for regenerative medicine.
Keywords: Growth factor; Mesenchymal stem cells (MSCs); Metabolite; Proteome; Regenerative medicine; Stemness maintenance.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Zhou Y, Fan W, Prasadam I, Crawford R, Xiao Y. Implantation of osteogenic differentiated donor mesenchymal stem cells causes recruitment of host cells. J Tissue Eng Regen Med. 2015;9:118–126. - PubMed
-
- Liesveld JL, Sharma N, Aljitawi OS. Stem cell homing: from physiology to therapeutics. Stem Cells. 2020;38:1241-53. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

