One year of SARS-CoV-2 evolution
- PMID: 33676588
- PMCID: PMC7903908
- DOI: 10.1016/j.chom.2021.02.017
One year of SARS-CoV-2 evolution
Abstract
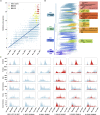
Since the outbreak of SARS-CoV-2, the etiologic agent of the COVID-19 pandemic, the viral genome has acquired numerous mutations with the potential to increase transmission. One year after its emergence, we now further analyze emergent SARS-CoV-2 genome sequences in an effort to understand the evolution of this virus.
Copyright © 2021. Published by Elsevier Inc.
Figures

References
-
- Bošnjak B., Stein S.C., Willenzon S., Cordes A.K., Puppe W., Bernhardt G., Ravens I., Ritter C., Schultze-Florey C.R., Gödecke N., et al. Low serum neutralizing anti-SARS-CoV-2 S antibody levels in mildly affected COVID-19 convalescent patients revealed by two different detection methods. Cell. Mol. Immunol. 2020 doi: 10.1038/s41423-020-00573-9. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

