Simplicity is the Ultimate Sophistication-Crosstalk of Post-translational Modifications on the RNA Polymerase II
- PMID: 33676925
- PMCID: PMC8184622
- DOI: 10.1016/j.jmb.2021.166912
Simplicity is the Ultimate Sophistication-Crosstalk of Post-translational Modifications on the RNA Polymerase II
Abstract
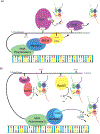
The highly conserved C-terminal domain (CTD) of the largest subunit of RNA polymerase II comprises a consensus heptad (Y1S2P3T4S5P6S7) repeated multiple times. Despite the simplicity of its sequence, the essential CTD domain orchestrates eukaryotic transcription and co-transcriptional processes, including transcription initiation, elongation, and termination, and mRNA processing. These distinct facets of the transcription cycle rely on specific post-translational modifications (PTM) of the CTD, in which five out of the seven residues in the heptad repeat are subject to phosphorylation. A hypothesis termed the "CTD code" has been proposed in which these PTMs and their combinations generate a sophisticated landscape for spatiotemporal recruitment of transcription regulators to Pol II. In this review, we summarize the recent experimental evidence understanding the biological role of the CTD, implicating a context-dependent theme that significantly enhances the ability of accurate transcription by RNA polymerase II. Furthermore, feedback communication between the CTD and histone modifications coordinates chromatin states with RNA polymerase II-mediated transcription, ensuring the effective and accurate conversion of information into cellular responses.
Keywords: RNA polymerase II; crosstalk; histone; phosphorylation; transcription.
Copyright © 2021 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



Similar articles
-
The RNA polymerase II CTD coordinates transcription and RNA processing.Genes Dev. 2012 Oct 1;26(19):2119-37. doi: 10.1101/gad.200303.112. Genes Dev. 2012. PMID: 23028141 Free PMC article. Review.
-
Removing quote marks from the RNA polymerase II CTD 'code'.Biosystems. 2021 Sep;207:104468. doi: 10.1016/j.biosystems.2021.104468. Epub 2021 Jun 30. Biosystems. 2021. PMID: 34216714
-
Transcription by RNA polymerase II and the CTD-chromatin crosstalk.Biochem Biophys Res Commun. 2022 Apr 9;599:81-86. doi: 10.1016/j.bbrc.2022.02.039. Epub 2022 Feb 11. Biochem Biophys Res Commun. 2022. PMID: 35176629 Review.
-
Phospho-site mutants of the RNA Polymerase II C-terminal domain alter subtelomeric gene expression and chromatin modification state in fission yeast.Nucleic Acids Res. 2016 Nov 2;44(19):9180-9189. doi: 10.1093/nar/gkw603. Epub 2016 Jul 8. Nucleic Acids Res. 2016. PMID: 27402158 Free PMC article.
-
Cracking the RNA polymerase II CTD code.Trends Genet. 2008 Jun;24(6):280-8. doi: 10.1016/j.tig.2008.03.008. Epub 2008 May 3. Trends Genet. 2008. PMID: 18457900 Review.
Cited by
-
Genome-Wide Regulations of the Preinitiation Complex Formation and Elongating RNA Polymerase II by an E3 Ubiquitin Ligase, San1.Mol Cell Biol. 2022 Jan 20;42(1):e0036821. doi: 10.1128/MCB.00368-21. Epub 2021 Oct 18. Mol Cell Biol. 2022. PMID: 34661445 Free PMC article.
-
Live imaging of transcription sites using an elongating RNA polymerase II-specific probe.J Cell Biol. 2022 Feb 7;221(2):e202104134. doi: 10.1083/jcb.202104134. Epub 2021 Dec 2. J Cell Biol. 2022. PMID: 34854870 Free PMC article.
-
Variation of C-terminal domain governs RNA polymerase II genomic locations and alternative splicing in eukaryotic transcription.bioRxiv [Preprint]. 2024 Jan 2:2024.01.01.573828. doi: 10.1101/2024.01.01.573828. bioRxiv. 2024. Update in: Nat Commun. 2024 Sep 12;15(1):7985. doi: 10.1038/s41467-024-52391-6. PMID: 38260389 Free PMC article. Updated. Preprint.
-
The cell biology of HIV-1 latency and rebound.Retrovirology. 2024 Apr 5;21(1):6. doi: 10.1186/s12977-024-00639-w. Retrovirology. 2024. PMID: 38580979 Free PMC article. Review.
-
RPRD1B's direct interaction with phosphorylated RNA polymerase II regulates polyadenylation of cell cycle genes and drives cancer progression.RSC Chem Biol. 2025 Jan 22;6(3):423-437. doi: 10.1039/d4cb00212a. eCollection 2025 Mar 5. RSC Chem Biol. 2025. PMID: 39886382 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

