Granulosa cell genes that regulate ovarian follicle development beyond the antral stage: The role of estrogen receptor β
- PMID: 33676987
- PMCID: PMC8916094
- DOI: 10.1016/j.mce.2021.111212
Granulosa cell genes that regulate ovarian follicle development beyond the antral stage: The role of estrogen receptor β
Abstract
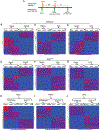
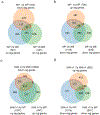
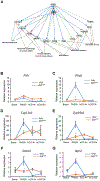
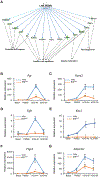
Follicle development beyond the preantral stage is dependent on gonadotropins. FSH signaling is crucial for the advancement of preantral follicles to the antral stage, and LH signaling is essential for further maturation of preovulatory follicles. Estrogen is intricately tied to gonadotropin signaling during the advanced stages of folliculogenesis. We observed that Erβnull ovarian follicles fail to develop beyond the antral stage, even after exogenous gonadotropin stimulation. As ERβ is primarily expressed in the granulosa cells (GCs), we explored the gonadotropin-regulated GC genes that induce maturation of antral follicles. Synchronized follicle development was induced by administration of exogenous gonadotropins to wildtype 4-wk-old female rats. The GC transcriptome was analyzed via RNA-sequencing before and after gonadotropin stimulation. An Erβnull mutant model that fails to show follicle maturation was also included in order to identify the ERβ-regulated genes involved at this step. We observed that specific groups of genes were differentially expressed in response to PMSG or hCG administration in wildtype rats. While some of the PMSG or hCG-induced genes showed a similar expression pattern in Erβnull GCs, a subset of PMSG- or hCG-induced genes showed a differential expression pattern in Erβnull GCs. These latter ERβ-regulated genes included previously known FSH or LH target genes including Lhcgr, Cyp11a1, Cyp19a1, Pgr, Runx2, Egfr, Kiss1, and Ptgs2, which are involved in follicle development, oocyte maturation, and ovulation. We also identified novel ERβ-regulated genes including Jaml, Galnt6, Znf750, Dusp9, Wnt16, and Mageb16 that failed to respond to gonadotropin stimulation in Erβnull GCs. Our findings indicate that the gonadotropin-induced spatiotemporal pattern of gene expression is essential for ovarian follicle maturation beyond the antral stage. However, expression of a subset of those gonadotropin-induced genes is dependent on transcriptional regulation by ERβ.
Keywords: ERβ; Follicle maturation; Gonadotropins; Granulosa cells; Transcriptome analyses.
Copyright © 2021 Elsevier B.V. All rights reserved.
Conflict of interest statement
Figures


Similar articles
-
ERβ Regulation of Gonadotropin Responses during Folliculogenesis.Int J Mol Sci. 2021 Sep 26;22(19):10348. doi: 10.3390/ijms221910348. Int J Mol Sci. 2021. PMID: 34638689 Free PMC article. Review.
-
The absence of ER-β results in altered gene expression in ovarian granulosa cells isolated from in vivo preovulatory follicles.Endocrinology. 2013 Jun;154(6):2174-87. doi: 10.1210/en.2012-2256. Epub 2013 Apr 11. Endocrinology. 2013. PMID: 23580569 Free PMC article.
-
In vitro growth and ovulation of follicles from ovaries of estrogen receptor (ER){alpha} and ER{beta} null mice indicate a role for ER{beta} in follicular maturation.Endocrinology. 2005 Jun;146(6):2817-26. doi: 10.1210/en.2004-1108. Epub 2005 Feb 24. Endocrinology. 2005. PMID: 15731357
-
Insufficient luteinizing hormone-induced intracellular signaling disrupts ovulation in preovulatory follicles lacking estrogen receptor-{beta}.Endocrinology. 2010 Jun;151(6):2826-34. doi: 10.1210/en.2009-1446. Epub 2010 Apr 8. Endocrinology. 2010. PMID: 20378682 Free PMC article.
-
Review: Roles of follicle-stimulating hormone in preantral folliculogenesis of domestic animals: what can we learn from model species and where do we go from here?Animal. 2023 May;17 Suppl 1:100743. doi: 10.1016/j.animal.2023.100743. Animal. 2023. PMID: 37567683 Review.
Cited by
-
Melatonin mitigates PGF-induced apoptosis during luteal regression in heat-exposed rats.Anim Reprod. 2025 Jun 30;22(2):e20240122. doi: 10.1590/1984-3143-AR2024-0122. eCollection 2025. Anim Reprod. 2025. PMID: 40599700 Free PMC article.
-
Ovarian ERβ cistrome and transcriptome reveal chromatin interaction with LRH-1.BMC Biol. 2023 Nov 29;21(1):277. doi: 10.1186/s12915-023-01773-1. BMC Biol. 2023. PMID: 38031019 Free PMC article.
-
Dynamic changes in gene expression of growing nonhuman primate antral follicles.Physiol Genomics. 2024 Nov 1;56(11):764-775. doi: 10.1152/physiolgenomics.00023.2024. Epub 2024 Sep 23. Physiol Genomics. 2024. PMID: 39311840
-
ERβ Regulation of Indian Hedgehog Expression in the First Wave of Ovarian Follicles.Cells. 2024 Apr 6;13(7):644. doi: 10.3390/cells13070644. Cells. 2024. PMID: 38607081 Free PMC article.
-
Altered Expression of Epigenetic and Transcriptional Regulators in ERβ Knockout Rat Ovaries During Postnatal Development.Int J Mol Sci. 2025 Jan 17;26(2):760. doi: 10.3390/ijms26020760. Int J Mol Sci. 2025. PMID: 39859473 Free PMC article.
References
-
- Bédard J, Brûlé S, Price CA, Silversides DW, and Lussier JG. 2003. Serine protease inhibitor-E2 (SERPINE2) is differentially expressed in granulosa cells of dominant follicle in cattle. Molecular reproduction and development. 64:152–165. - PubMed
-
- Brown HM, Dunning KR, Robker RL, Boerboom D, Pritchard M, Lane M, and Russell DL. 2010. ADAMTS1 cleavage of versican mediates essential structural remodeling of the ovarian follicle and cumulus-oocyte matrix during ovulation in mice. Biology of reproduction. 83:549–557. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

