Quantitative Proteomics and Phosphoproteomics Support a Role for Mut9-Like Kinases in Multiple Metabolic and Signaling Pathways in Arabidopsis
- PMID: 33677124
- PMCID: PMC8066427
- DOI: 10.1016/j.mcpro.2021.100063
Quantitative Proteomics and Phosphoproteomics Support a Role for Mut9-Like Kinases in Multiple Metabolic and Signaling Pathways in Arabidopsis
Abstract
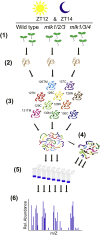
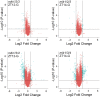
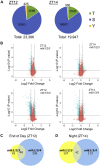
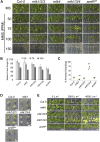
Protein phosphorylation is one of the most prevalent posttranslational modifications found in eukaryotic systems. It serves as a key molecular mechanism that regulates protein function in response to environmental stimuli. The Mut9-like kinases (MLKs) are a plant-specific family of Ser/Thr kinases linked to light, circadian, and abiotic stress signaling. Here we use quantitative phosphoproteomics in conjunction with global proteomic analysis to explore the role of the MLKs in daily protein dynamics. Proteins involved in light, circadian, and hormone signaling, as well as several chromatin-modifying enzymes and DNA damage response factors, were found to have altered phosphorylation profiles in the absence of MLK family kinases. In addition to altered phosphorylation levels, mlk mutant seedlings have an increase in glucosinolate metabolism enzymes. Subsequently, we show that a functional consequence of the changes to the proteome and phosphoproteome in mlk mutant plants is elevated glucosinolate accumulation and increased sensitivity to DNA damaging agents. Combined with previous reports, this work supports the involvement of MLKs in a diverse set of stress responses and developmental processes, suggesting that the MLKs serve as key regulators linking environmental inputs to developmental outputs.
Keywords: DNA damage; arabidopsis; circadian; kinase; plants; quantitative phosphoproteomics; quantitative proteomics.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare no competing interests.
Figures






References
-
- Mergner J., Frejno M., List M., Papacek M., Chen X., Chaudhary A., Samaras P., Richter S., Shikata H., Messerer M., Lang D., Altmann S., Cyprys P., Zolg D.P., Mathieson T. Mass-spectrometry-based draft of the Arabidopsis proteome. Nature. 2020;579:409–414. - PubMed
-
- Silva-Sanchez C., Li H., Chen S. Recent advances and challenges in plant phosphoproteomics. Proteomics. 2015;15:1127–1141. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

