An expanded auxin-inducible degron toolkit for Caenorhabditis elegans
- PMID: 33677541
- PMCID: PMC8045686
- DOI: 10.1093/genetics/iyab006
An expanded auxin-inducible degron toolkit for Caenorhabditis elegans
Abstract
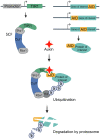
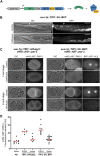
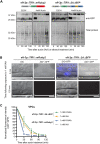
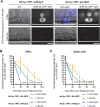
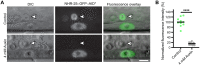
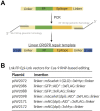
The auxin-inducible degron (AID) system has emerged as a powerful tool to conditionally deplete proteins in a range of organisms and cell types. Here, we describe a toolkit to augment the use of the AID system in Caenorhabditis elegans. We have generated a set of single-copy, tissue-specific (germline, intestine, neuron, muscle, pharynx, hypodermis, seam cell, anchor cell) and pan-somatic TIR1-expressing strains carrying a co-expressed blue fluorescent reporter to enable use of both red and green channels in experiments. These transgenes are inserted into commonly used, well-characterized genetic loci. We confirmed that our TIR1-expressing strains produce the expected depletion phenotype for several nuclear and cytoplasmic AID-tagged endogenous substrates. We have also constructed a set of plasmids for constructing repair templates to generate fluorescent protein::AID fusions through CRISPR/Cas9-mediated genome editing. These plasmids are compatible with commonly used genome editing approaches in the C. elegans community (Gibson or SapTrap assembly of plasmid repair templates or PCR-derived linear repair templates). Together these reagents will complement existing TIR1 strains and facilitate rapid and high-throughput fluorescent protein::AID tagging of genes. This battery of new TIR1-expressing strains and modular, efficient cloning vectors serves as a platform for straightforward assembly of CRISPR/Cas9 repair templates for conditional protein depletion.
Keywords: C. elegans; AID system; CRISPR/Cas9; SapTrap; Transport Inhibitor Response 1; self-excising cassette.
© The Author(s) 2021. Published by Oxford University Press on behalf of Genetics Society of America.
Figures



References
-
- Breunig JJ, Levy R, Antonuk CD, Molina J, Dutra-Clarke M, et al. 2015. Ets factors regulate neural stem cell depletion and gliogenesis in Ras pathway glioma. Cell Rep. 12:258–271. http://doi:10.1016/j.celrep.2015.06.012 - DOI - PubMed
-
- Brown KM, Long S, Sibley LD.. 2017. Plasma membrane association by N-acylation governs pkg function in Toxoplasma gondii. mBio. 8:e00375–17. http://doi:10.1128/mBio.00375-17 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P40 OD010440/OD/NIH HHS/United States
- T32 GM008444/GM/NIGMS NIH HHS/United States
- R00 GM115964/GM/NIGMS NIH HHS/United States
- K99 GM107345/GM/NIGMS NIH HHS/United States
- R00 GM107345/GM/NIGMS NIH HHS/United States
- F32 GM131577/GM/NIGMS NIH HHS/United States
- R01 GM121625/GM/NIGMS NIH HHS/United States
- R01 GM138701/GM/NIGMS NIH HHS/United States
- F31 HD100091/HD/NICHD NIH HHS/United States
- R35 GM134838/GM/NIGMS NIH HHS/United States
- T32 GM133391/GM/NIGMS NIH HHS/United States
- R01 GM121597/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

