Mobile Carbapenemase Genes in Pseudomonas aeruginosa
- PMID: 33679638
- PMCID: PMC7930500
- DOI: 10.3389/fmicb.2021.614058
Mobile Carbapenemase Genes in Pseudomonas aeruginosa
Abstract
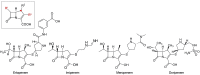
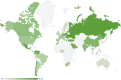
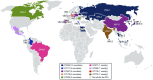
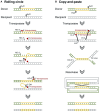
Carbapenem-resistant Pseudomonas aeruginosa is one of the major concerns in clinical settings impelling a great challenge to antimicrobial therapy for patients with infections caused by the pathogen. While membrane permeability, together with derepression of the intrinsic beta-lactamase gene, is the global prevailing mechanism of carbapenem resistance in P. aeruginosa, the acquired genes for carbapenemases need special attention because horizontal gene transfer through mobile genetic elements, such as integrons, transposons, plasmids, and integrative and conjugative elements, could accelerate the dissemination of the carbapenem-resistant P. aeruginosa. This review aimed to illustrate epidemiologically the carbapenem resistance in P. aeruginosa, including the resistance rates worldwide and the carbapenemase-encoding genes along with the mobile genetic elements responsible for the horizontal dissemination of the drug resistance determinants. Moreover, the modular mobile elements including the carbapenemase-encoding gene, also known as the P. aeruginosa resistance islands, are scrutinized mostly for their structures.
Keywords: Pseudomonas aeruginosa; carbapenem resistance; carbapenemase; genomic islands; mobile genetic elements; molecular epidemiology.
Copyright © 2021 Yoon and Jeong.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



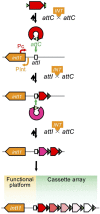
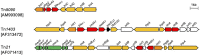
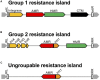
References
-
- Abril D., Marquez-Ortiz R. A., Castro-Cardozo B., Moncayo-Ortiz J. I., Olarte Escobar N. M., Corredor Rozo Z. L., et al. (2019). Genome plasticity favours double chromosomal Tn4401b-blaKPC-2 transposon insertion in the Pseudomonas aeruginosa ST235 clone. BMC Microbiol. 19:45. 10.1186/s12866-019-1418-6 - DOI - PMC - PubMed
-
- Al-Agamy M. H., Shibl A. M., Tawfik A. F., Radwan H. H. (2009). High prevalence of metallo-beta-lactamase-producing Pseudomonas aeruginosa from Saudi Arabia. J. Chemother. 21 461–462. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

