Vibrio natriegens as a pET-Compatible Expression Host Complementary to Escherichia coli
- PMID: 33679648
- PMCID: PMC7933001
- DOI: 10.3389/fmicb.2021.627181
Vibrio natriegens as a pET-Compatible Expression Host Complementary to Escherichia coli
Abstract
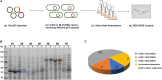
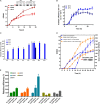
Efficient and novel recombinant protein expression systems can further reduce the production cost of enzymes. Vibrio natriegens is the fastest growing free-living bacterium with a doubling time of less than 10 min, which makes it highly attractive as a protein expression host. Here, 196 pET plasmids with different genes of interest (GOIs) were electroporated into the V. natriegens strain VnDX, which carries an integrated T7 RNA polymerase expression cassette. As a result, 65 and 75% of the tested GOIs obtained soluble expression in V. natriegens and Escherichia coli, respectively, 20 GOIs of which showed better expression in the former. Furthermore, we have adapted a consensus "what to try first" protocol for V. natriegens based on Terrific Broth medium. Six sampled GOIs encoding biocatalysts enzymes thus achieved 50-128% higher catalytic efficiency under the optimized expression conditions. Our study demonstrated V. natriegens as a pET-compatible expression host with a spectrum of highly expressed GOIs distinct from E. coli and an easy-to-use consensus protocol, solving the problem that some GOIs cannot be expressed well in E. coli.
Keywords: Vibrio natriegens; fermentation optimization; pET expression system; recombinant protein; synthetic biology.
Copyright © 2021 Xu, Dong, Wu, Tao, Yang, Wu, Jiang, Yang and Yang.
Conflict of interest statement
RT was employed by company Huzhou Yisheng Biotechnology Co., Ltd. YJ was employed by company Shanghai Taoyusheng Biotechnology Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Broedel S. J., Papciak S., Jones W. (2001). The selection of optimum media formulations for improved expression of recombinant proteins in E. coli. Technic. Bull. 2.
-
- Bucher T., Krell H., Lusch G. (1974). Molar extinction coefficients of NADH and NADPH at Hg spectral lines. Z. Klin. Chem. Klin. Biochem. 12 239–240. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

