Metabolic Potential, Ecology and Presence of Associated Bacteria Is Reflected in Genomic Diversity of Mucoromycotina
- PMID: 33679672
- PMCID: PMC7928374
- DOI: 10.3389/fmicb.2021.636986
Metabolic Potential, Ecology and Presence of Associated Bacteria Is Reflected in Genomic Diversity of Mucoromycotina
Abstract
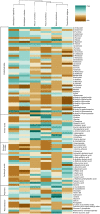
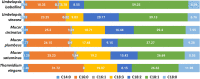
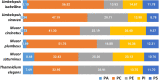
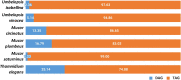
Mucoromycotina are often considered mainly in pathogenic context but their biology remains understudied. We describe the genomes of six Mucoromycotina fungi representing distant saprotrophic lineages within the subphylum (i.e., Umbelopsidales and Mucorales). We selected two Umbelopsis isolates from soil (i.e., U. isabellina, U. vinacea), two soil-derived Mucor isolates (i.e., M. circinatus, M. plumbeus), and two Mucorales representatives with extended proteolytic activity (i.e., Thamnidium elegans and Mucor saturninus). We complement computational genome annotation with experimental characteristics of their digestive capabilities, cell wall carbohydrate composition, and extensive total lipid profiles. These traits inferred from genome composition, e.g., in terms of identified encoded enzymes, are in accordance with experimental results. Finally, we link the presence of associated bacteria with observed characteristics. Thamnidium elegans genome harbors an additional, complete genome of an associated bacterium classified to Paenibacillus sp. This fungus displays multiple altered traits compared to the remaining isolates, regardless of their evolutionary distance. For instance, it has expanded carbon assimilation capabilities, e.g., efficiently degrades carboxylic acids, and has a higher diacylglycerol:triacylglycerol ratio and skewed phospholipid composition which suggests a more rigid cellular membrane. The bacterium can complement the host enzymatic capabilities, alter the fungal metabolism, cell membrane composition but does not change the composition of the cell wall of the fungus. Comparison of early-diverging Umbelopsidales with evolutionary younger Mucorales points at several subtle differences particularly in their carbon source preferences and encoded carbohydrate repertoire. Nevertheless, all tested Mucoromycotina share features including the ability to produce 18:3 gamma-linoleic acid, use TAG as the storage lipid and have fucose as a cell wall component.
Keywords: Mucorales; Paenibacillus; Umbelopsidales; carbon source usage; cell wall carbohydrates; comparative genomics; lipid profile.
Copyright © 2021 Muszewska, Okrasińska, Steczkiewicz, Drgas, Orłowska, Perlińska-Lenart, Aleksandrzak-Piekarczyk, Szatraj, Zielenkiewicz, Piłsyk, Malc, Mieczkowski, Kruszewska, Bernat and Pawłowska.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Carbon assimilation profiles of mucoralean fungi show their metabolic versatility.Sci Rep. 2019 Aug 14;9(1):11864. doi: 10.1038/s41598-019-48296-w. Sci Rep. 2019. PMID: 31413281 Free PMC article.
-
Fucose as a nutrient ligand for Dikarya and a building block of early diverging lineages.IMA Fungus. 2023 Sep 5;14(1):17. doi: 10.1186/s43008-023-00123-8. IMA Fungus. 2023. PMID: 37670396 Free PMC article.
-
Preliminary studies on the evolution of carbon assimilation abilities within Mucorales.Fungal Biol. 2016 May;120(5):752-63. doi: 10.1016/j.funbio.2016.02.004. Epub 2016 Feb 18. Fungal Biol. 2016. PMID: 27109371
-
Outbreaks of Mucorales and the Species Involved.Mycopathologia. 2020 Oct;185(5):765-781. doi: 10.1007/s11046-019-00403-1. Epub 2019 Nov 16. Mycopathologia. 2020. PMID: 31734800 Review.
-
Iron Assimilation during Emerging Infections Caused by Opportunistic Fungi with emphasis on Mucorales and the Development of Antifungal Resistance.Genes (Basel). 2020 Oct 30;11(11):1296. doi: 10.3390/genes11111296. Genes (Basel). 2020. PMID: 33143139 Free PMC article. Review.
Cited by
-
Sequencing the Genomes of the First Terrestrial Fungal Lineages: What Have We Learned?Microorganisms. 2023 Jul 18;11(7):1830. doi: 10.3390/microorganisms11071830. Microorganisms. 2023. PMID: 37513002 Free PMC article. Review.
-
The endohyphal microbiome: current progress and challenges for scaling down integrative multi-omic microbiome research.Microbiome. 2023 Aug 26;11(1):192. doi: 10.1186/s40168-023-01634-7. Microbiome. 2023. PMID: 37626434 Free PMC article.
-
Detecting and characterizing new endofungal bacteria in new hosts: Pandoraea sputorum and Mycetohabitans endofungorum in Rhizopus arrhizus.Front Microbiol. 2024 Feb 29;15:1346252. doi: 10.3389/fmicb.2024.1346252. eCollection 2024. Front Microbiol. 2024. PMID: 38486702 Free PMC article.
-
Ant's Nest as a microenvironment: Distinct Mucoromycota (Fungi) community of the red wood ants' (Formica polyctena) mounds.Ecol Evol. 2024 Oct 9;14(10):e70333. doi: 10.1002/ece3.70333. eCollection 2024 Oct. Ecol Evol. 2024. PMID: 39385841 Free PMC article.
-
Compatible traits of oleaginous Mucoromycota fungi for lignocellulose-based simultaneous saccharification and fermentation.Biotechnol Biofuels Bioprod. 2025 Feb 24;18(1):24. doi: 10.1186/s13068-025-02621-w. Biotechnol Biofuels Bioprod. 2025. PMID: 39994750 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

