Inflammation and Antiviral Immune Response Associated With Severe Progression of COVID-19
- PMID: 33679778
- PMCID: PMC7930228
- DOI: 10.3389/fimmu.2021.631226
Inflammation and Antiviral Immune Response Associated With Severe Progression of COVID-19
Abstract
Coronavirus disease-2019 (COVID-19) is a novel respiratory disease induced by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). It remains poorly understood how the host immune system responds to the infection during disease progression. We applied microarray analysis of the whole genome transcriptome to peripheral blood mononuclear cells (PBMCs) taken from severe and mild COVID-19 patients as well as healthy controls. Functional enrichment analysis of genes associated with COVID-19 severity indicated that disease progression is featured by overactivation of myeloid cells and deficient T cell function. The upregulation of TLR6 and MMP9, which promote the neutrophils-mediated inflammatory response, and the downregulation of SKAP1 and LAG3, which regulate T cells function, were associated with disease severity. Importantly, the regulation of these four genes was absent in patients with influenza A (H1N1). And compared with stimulation with hemagglutinin (HA) of H1N1 virus, the regulation pattern of these genes was unique in PBMCs response to Spike protein of SARS-CoV-2 ex vivo. Our data also suggested that severe SARS-CoV-2 infection largely silenced the response of type I interferons (IFNs) and altered the proportion of immune cells, providing a potential mechanism for the hypercytokinemia. This study indicates that SARS-CoV-2 infection impairs inflammatory and immune signatures in patients, especially those at severe stage. The potential mechanisms underpinning severe COVID-19 progression include overactive myeloid cells, impaired function of T cells, and inadequate induction of type I IFNs signaling.
Keywords: COVID-19; SARS-CoV-2; T cells; immune response; inflammation; interferons; myeloid cells; transcriptome.
Copyright © 2021 Zhang, Meng, Wang, Zhang, Chen, Sheng, Qiu, Diao and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

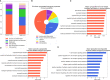

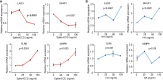
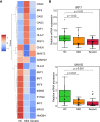
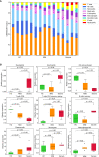
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

