Applying Molecular Phenotyping Tools to Explore Sugarcane Carbon Potential
- PMID: 33679852
- PMCID: PMC7935522
- DOI: 10.3389/fpls.2021.637166
Applying Molecular Phenotyping Tools to Explore Sugarcane Carbon Potential
Abstract
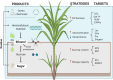
Sugarcane (Saccharum spp.), a C4 grass, has a peculiar feature: it accumulates, gradient-wise, large amounts of carbon (C) as sucrose in its culms through a complex pathway. Apart from being a sustainable crop concerning C efficiency and bioenergetic yield per hectare, sugarcane is used as feedstock for producing ethanol, sugar, high-value compounds, and products (e.g., polymers and succinate), and bioelectricity, earning the title of the world's leading biomass crop. Commercial cultivars, hybrids bearing high levels of polyploidy, and aneuploidy, are selected from a large number of crosses among suitable parental genotypes followed by the cloning of superior individuals among the progeny. Traditionally, these classical breeding strategies have been favoring the selection of cultivars with high sucrose content and resistance to environmental stresses. A current paradigm change in sugarcane breeding programs aims to alter the balance of C partitioning as a means to provide more plasticity in the sustainable use of this biomass for metabolic engineering and green chemistry. The recently available sugarcane genetic assemblies powered by data science provide exciting perspectives to increase biomass, as the current sugarcane yield is roughly 20% of its predicted potential. Nowadays, several molecular phenotyping tools can be applied to meet the predicted sugarcane C potential, mainly targeting two competing pathways: sucrose production/storage and biomass accumulation. Here we discuss how molecular phenotyping can be a powerful tool to assist breeding programs and which strategies could be adopted depending on the desired final products. We also tackle the advances in genetic markers and mapping as well as how functional genomics and genetic transformation might be able to improve yield and saccharification rates. Finally, we review how "omics" advances are promising to speed up plant breeding and reach the unexplored potential of sugarcane in terms of sucrose and biomass production.
Keywords: biomass; carbon; molecular phenotyping; sucrose; sugarcane.
Copyright © 2021 Calderan-Rodrigues, de Barros Dantas, Cheavegatti Gianotto and Caldana.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Aljanabi S. M., Parmessur Y., Kross H., Dhayan S., Saumtally S., Ramdoyal K., et al. (2006). Identification of a major Quantitative Trait Locus (QTL) for yellow spot (Mycovellosiella koepkei) disease resistance in sugarcane. Mol. Breed. 19 1–14. 10.1007/s11032-006-9008-3 - DOI
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

