Circular RNAs Sparkle in the Diagnosis and Theranostics of Hepatocellular Carcinoma
- PMID: 33679871
- PMCID: PMC7930616
- DOI: 10.3389/fgene.2020.628655
Circular RNAs Sparkle in the Diagnosis and Theranostics of Hepatocellular Carcinoma
Abstract
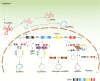
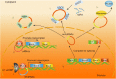
Exonic circular RNAs (circRNAs) are a novel subgroup of non-coding RNAs, which are generated by a back-splicing mechanism of the exons or introns. Unlike the linear RNA, circRNA forms a covalently closed loop, and it normally appears more abundant than the linear products of its host gene. Due to the relatively high specificity and stability of circular RNAs in tissues and body fluid, circular RNAs have attracted widely scientific interest for its potential application in cancer diagnosis and as a guide for preclinical therapy, especially for hard-to-treat cancers with high heterogeneity, such as hepatocellular carcinoma (HCC). Thus, we summarize the updated knowledge of circular RNAs, including the mechanism of the generation of endogenous circular RNAs and their regulatory, diagnostic, and therapeutic roles in HCC.
Keywords: HCC; biomarker; circRNA; hepatocellular carcinoma; therapy.
Copyright © 2021 Wang, Wu, Xie and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

