Integrated Bioinformatics Analysis Exhibits Pivotal Exercise-Induced Genes and Corresponding Pathways in Malignant Melanoma
- PMID: 33679872
- PMCID: PMC7930906
- DOI: 10.3389/fgene.2020.637320
Integrated Bioinformatics Analysis Exhibits Pivotal Exercise-Induced Genes and Corresponding Pathways in Malignant Melanoma
Abstract
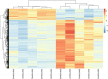
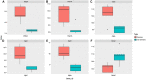
Malignant melanoma represents a sort of neoplasm deriving from melanocytes or cells developing from melanocytes. The balance of energy and energy-associated body composition and body mass index could be altered by exercise, thereby directly affecting the microenvironment of neoplasm. However, few studies have examined the mechanism of genes induced by exercise and the pathways involved in melanoma. This study used three separate datasets to perform comprehensive bioinformatics analysis and then screened the probable genes and pathways in the process of exercise-promoted melanoma. In total, 1,627 differentially expressed genes (DEGs) induced by exercise were recognized. All selected genes were largely enriched in NF-kappa B, Chemokine signaling pathways, and the immune response after gene set enrichment analysis. The protein-protein interaction network was applied to excavate DEGs and identified the most relevant and pivotal genes. The top 6 hub genes (Itgb2, Wdfy4, Itgam, Cybb, Mmp2, and Parp14) were identified, and importantly, 5 hub genes (Itgb2, Wdfy4, Itgam, Cybb, and Parp14) were related to weak disease-free survival and overall survival (OS). In conclusion, our findings demonstrate the prognostic value of exercise-induced genes and uncovered the pathways of these genes in melanoma, implying that these genes might act as prognostic biomarkers for melanoma.
Keywords: Disease-free survival; exercise; integrated bioinformatics analysis; malignant melanoma; prognosis.
Copyright © 2021 Zhu, Hao, Zhang, Qiu, Xuan and Ye.
Conflict of interest statement
SH was employed by company Shuangwu Information Technical Company Ltd, China. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Azijli K., Stelloo E., Peters G. J., Van Den Eertwegh A. J. M. (2014). New developments in the treatment of metastatic melanoma: immune checkpoint inhibitors and targeted therapies. Anticancer Res. 34 1493–1505. - PubMed
-
- Azzola M. F., Shaw H. M., Thompson J. F., Soong S. J., Scolyer R. A., Watson G. F., et al. (2003). Tumor mitotic rate is a more powerful prognostic indicator than ulceration in patients with primary cutaneous melanoma: an analysis of 3661 patients from a single center. Cancer 97 1488–1498. 10.1002/cncr.11196 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

