Plant cell wall architecture guided design of CBM3-GH11 chimeras with enhanced xylanase activity using a tandem repeat left-handed β-3-prism scaffold
- PMID: 33680354
- PMCID: PMC7890094
- DOI: 10.1016/j.csbj.2021.01.011
Plant cell wall architecture guided design of CBM3-GH11 chimeras with enhanced xylanase activity using a tandem repeat left-handed β-3-prism scaffold
Abstract
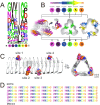
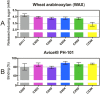
Effective use of plant biomass as an abundant and renewable feedstock for biofuel production and biorefinery requires efficient enzymatic mobilization of cell wall polymers. Knowledge of plant cell wall composition and architecture has been exploited to develop novel multifunctional enzymes with improved activity against lignocellulose, where a left-handed β-3-prism synthetic scaffold (BeSS) was designed for insertion of multiple protein domains at the prism vertices. This allowed construction of a series of chimeras fusing variable numbers of a GH11 β-endo-1,4-xylanase and the CipA-CBM3 with defined distances and constrained relative orientations between catalytic domains. The cellulose binding and endoxylanase activities of all chimeras were maintained. Activity against lignocellulose substrates revealed a rapid 1.6- to 3-fold increase in total reducing saccharide release and increased levels of all major oligosaccharides as measured by polysaccharide analysis using carbohydrate gel electrophoresis (PACE). A construct with CBM3 and GH11 domains inserted in the same prism vertex showed highest activity, demonstrating interdomain geometry rather than number of catalytic sites is important for optimized chimera design. These results confirm that the BeSS concept is robust and can be successfully applied to the construction of multifunctional chimeras, which expands the possibilities for knowledge-based protein design.
Keywords: Carbohydrate-binding module; Endoxylanase; Lignocellulose hydrolysis; Multifunctional protein; Protein design; Protein engineering; Synthetic biology.
© 2021 The Authors.
Conflict of interest statement
The authors declare no conflict no interest associated with the work described in this manuscript.
Figures



Similar articles
-
The family 22 carbohydrate-binding module of bifunctional xylanase/β-glucanase Xyn10E from Paenibacillus curdlanolyticus B-6 has an important role in lignocellulose degradation.Enzyme Microb Technol. 2017 Jan;96:75-84. doi: 10.1016/j.enzmictec.2016.09.015. Epub 2016 Sep 26. Enzyme Microb Technol. 2017. PMID: 27871388
-
Functional Analysis of the Glucan Degradation Locus in Caldicellulosiruptor bescii Reveals Essential Roles of Component Glycoside Hydrolases in Plant Biomass Deconstruction.Appl Environ Microbiol. 2017 Dec 1;83(24):e01828-17. doi: 10.1128/AEM.01828-17. Print 2017 Dec 15. Appl Environ Microbiol. 2017. PMID: 28986379 Free PMC article.
-
Family 3 CBM improves the biochemical properties, substrate hydrolysis and coconut oil extraction by hemicellulolytic and holocellulolytic chimeras.Enzyme Microb Technol. 2024 Mar;174:110375. doi: 10.1016/j.enzmictec.2023.110375. Epub 2023 Dec 12. Enzyme Microb Technol. 2024. PMID: 38157781
-
Endo-1,4-β-xylanase-containing glycoside hydrolase families: characteristics, singularities and similarities.Biotechnol Adv. 2023 Jul-Aug;65:108148. doi: 10.1016/j.biotechadv.2023.108148. Epub 2023 Apr 7. Biotechnol Adv. 2023. PMID: 37030552 Review.
-
Engineering grass biomass for sustainable and enhanced bioethanol production.Planta. 2019 Aug;250(2):395-412. doi: 10.1007/s00425-019-03218-y. Epub 2019 Jun 24. Planta. 2019. PMID: 31236698 Review.
Cited by
-
Current challenges in designer cellulosome engineering.Appl Microbiol Biotechnol. 2023 May;107(9):2755-2770. doi: 10.1007/s00253-023-12474-8. Epub 2023 Mar 21. Appl Microbiol Biotechnol. 2023. PMID: 36941434 Review.
References
-
- Arai T., Biely P., Uhliariková I., Sato N., Makishima S., Mizuno M., Nozaki K., Kaneko S., Amano Y. Structural characterization of hemicellulose released from corn cob in continuous flow type hydrothermal reactor. J Biosci Bioeng. 2019;127(2):222–230. doi: 10.1016/j.jbiosc.2018.07.016. - DOI - PubMed
-
- Benhar I., Tamarkin A., Marash L., Berdichevsky Y., Yaron S., Shoham Y., Lamed R., Bayer E.A. Glycosyl Hydrolases for Biomass Conversion. American Chemical Society; 2000. Phage display of cellulose binding domains for biotechnological application; pp. 10–168. - DOI
-
- Boraston A.B., Bolam D.N., Gilbert H.J., Davies G.J. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J. 2004;382(3):769–781. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dop... - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
