Composition and distribution of fish environmental DNA in an Adirondack watershed
- PMID: 33680576
- PMCID: PMC7919543
- DOI: 10.7717/peerj.10539
Composition and distribution of fish environmental DNA in an Adirondack watershed
Abstract
Background: Environmental DNA (eDNA) surveys are appealing options for monitoring aquatic biodiversity. While factors affecting eDNA persistence, capture and amplification have been heavily studied, watershed-scale surveys of fish communities and our confidence in such need further exploration.
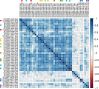
Methods: We characterized fish eDNA compositions using rapid, low-volume filtering with replicate and control samples scaled for a single Illumina MiSeq flow cell, using the mitochondrial 12S ribosomal RNA locus for taxonomic profiling. Our goals were to determine: (1) spatiotemporal variation in eDNA abundance, (2) the filtrate needed to achieve strong sequencing libraries, (3) the taxonomic resolution of 12S ribosomal sequences in the study environment, (4) the portion of the expected fish community detectable by 12S sequencing, (5) biases in species recovery, (6) correlations between eDNA compositions and catch per unit effort (CPUE) and (7) the extent that eDNA profiles reflect major watershed features. Our bioinformatic approach included (1) estimation of sequencing error from unambiguous mappings and simulation of taxonomic assignment error under various mapping criteria; (2) binning of species based on inferred assignment error rather than by taxonomic rank; and (3) visualization of mismatch distributions to facilitate discovery of distinct haplotypes attributed to the same reference. Our approach was implemented within the St. Regis River, NY, USA, which supports tribal and recreational fisheries and has been a target of restoration activities. We used a large record of St. Regis-specific observations to validate our assignments.
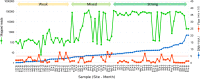
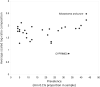
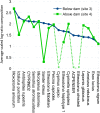
Results: We found that 300 mL drawn through 25-mm cellulose nitrate filters yielded greater than 5 ng/µL DNA at most sites in summer, which was an approximate threshold for generating strong sequencing libraries in our hands. Using inferred sequence error rates, we binned 12S references for 110 species on a state checklist into 85 single-species bins and seven multispecies bins. Of 48 bins observed by capture survey in the St. Regis, we detected eDNA consistent with 40, with an additional four detections flagged as potential contaminants. Sixteen unobserved species detected by eDNA ranged from plausible to implausible based on distributional data, whereas six observed species had no 12S reference sequence. Summed log-ratio compositions of eDNA-detected taxa correlated with log(CPUE) (Pearson's R = 0.655, P < 0.001). Shifts in eDNA composition of several taxa and a genotypic shift in channel catfish (Ictalurus punctatus) coincided with the Hogansburg Dam, NY, USA. In summary, a simple filtering apparatus operated by field crews without prior expertise gave useful summaries of eDNA composition with minimal evidence of field contamination. 12S sequencing achieved useful taxonomic resolution despite the short marker length, and data exploration with standard bioinformatic tools clarified taxonomic uncertainty and sources of error.
Keywords: Barcode sequencing; Computational biology; Dam removal; Environmental DNA; Fisheries restoration; Metagenetics; Mitochondrial 12S ribosomal RNA; New York state.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
Evaluation of Fish Species Detection in the Northwestern Pacific using eDNA Metabarcoding: A Mock Community Approach.Front Biosci (Schol Ed). 2025 Mar 24;17(1):26247. doi: 10.31083/FBS26247. Front Biosci (Schol Ed). 2025. PMID: 40150871
-
Integrating eDNA metabarcoding and simultaneous underwater visual surveys to describe complex fish communities in a marine biodiversity hotspot.Mol Ecol Resour. 2021 Jul;21(5):1558-1574. doi: 10.1111/1755-0998.13375. Epub 2021 Mar 31. Mol Ecol Resour. 2021. PMID: 33683812
-
Assessing vertebrate biodiversity in a kelp forest ecosystem using environmental DNA.Mol Ecol. 2016 Jan;25(2):527-41. doi: 10.1111/mec.13481. Epub 2015 Dec 24. Mol Ecol. 2016. PMID: 26586544 Free PMC article.
-
Standards for Methods Utilizing Environmental DNA for Detection of Fish Species.Genes (Basel). 2020 Mar 11;11(3):296. doi: 10.3390/genes11030296. Genes (Basel). 2020. PMID: 32168762 Free PMC article. Review.
-
Can we manage fisheries with the inherent uncertainty from eDNA?J Fish Biol. 2021 Feb;98(2):341-353. doi: 10.1111/jfb.14218. Epub 2019 Dec 19. J Fish Biol. 2021. PMID: 31769024 Review.
Cited by
-
Common methods in mitochondrial research (Review).Int J Mol Med. 2022 Oct;50(4):126. doi: 10.3892/ijmm.2022.5182. Epub 2022 Aug 25. Int J Mol Med. 2022. PMID: 36004457 Free PMC article.
-
Assessing arthropod diversity metrics derived from stream environmental DNA: spatiotemporal variation and paired comparisons with manual sampling.PeerJ. 2023 Mar 31;11:e15163. doi: 10.7717/peerj.15163. eCollection 2023. PeerJ. 2023. PMID: 37020852 Free PMC article.
References
-
- Andrew S. FastQC. 2020. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [10 August 2020]. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
-
- Barnes MA, Turner CR. The ecology of environmental DNA and implications for conservation genetics. Conservation Genetics. 2016;17(1):1–17. doi: 10.1007/s10592-015-0775-4. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

