MYCT1 Inhibits the Adhesion and Migration of Laryngeal Cancer Cells Potentially Through Repressing Collagen VI
- PMID: 33680912
- PMCID: PMC7931689
- DOI: 10.3389/fonc.2020.564733
MYCT1 Inhibits the Adhesion and Migration of Laryngeal Cancer Cells Potentially Through Repressing Collagen VI
Abstract
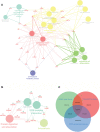
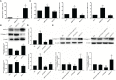
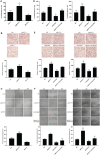
MYCT1, a target of c-Myc, inhibits laryngeal cancer cell migration, but the underlying mechanism remains unclear. In the study, we detected differentially expressed genes (DEGs) from laryngeal cancer cells transfected by MYCT1 using RNA-seq (GSE123275). DEGs from head and neck squamous cell carcinoma (HNSCC) were first screened by comparison of transcription data from the Gene Expression Omnibus (GSE6631) and the Cancer Genome Atlas (TCGA) datasets using weighted gene co-expression network analysis (WGCNA). GO and KEGG pathway analysis explained the functions of the DEGs. The DEGs overlapped between GSE6631and TCGA datasets were then compared with ours to find the key DEGs downstream of MYCT1 related to the adhesion and migration of laryngeal cancer cells. qRT-PCR and Western blot were applied to validate gene expression at mRNA and protein levels, respectively. Finally, the cell adhesion, migration, and wound healing assays were to check cell adhesion and migration abilities, respectively. As results, 39 overlapping genes were enriched in the GSE6631 and TCGA datasets, and most of them revealed adhesion function. Thirteen of 39 genes including COL6 members COL6A1, COL6A2, and COL6A3 were overlapped in GSE6631, TCGA, and GSE123275 datasets. Similar to our RNA-seq results, we confirmed that COL6 is a target of MYCT1 in laryngeal cancer cells. We also found that MYCT1 inhibited the adhesion and migration of laryngeal cancer cells via COL6. These indicate that COL6 is a potential target of MYCT1 and participates the adhesion and migration of laryngeal cancer cells, which provides an important clue for further study on how MYCT1 regulating COL6 in laryngeal cancer progression.
Keywords: Collagen Ⅵ; MYCT1; gene interaction network; laryngeal cancer; progression.
Copyright © 2021 Wang, Ding, Sun, Li and Fu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
MYCT1 inhibits the EMT and migration of laryngeal cancer cells via the SP1/miR-629-3p/ESRP2 pathway.Cell Signal. 2020 Oct;74:109709. doi: 10.1016/j.cellsig.2020.109709. Epub 2020 Jul 11. Cell Signal. 2020. PMID: 32659265
-
CREB promotes laryngeal cancer cell migration via MYCT1/NAT10 axis.Onco Targets Ther. 2018 Mar 8;11:1323-1331. doi: 10.2147/OTT.S156582. eCollection 2018. Onco Targets Ther. 2018. PMID: 29563811 Free PMC article.
-
YY1 directly suppresses MYCT1 leading to laryngeal tumorigenesis and progress.Cancer Med. 2017 Jun;6(6):1389-1398. doi: 10.1002/cam4.1073. Epub 2017 May 9. Cancer Med. 2017. PMID: 28485541 Free PMC article.
-
Identification of Potential Biomarkers and Survival Analysis for Head and Neck Squamous Cell Carcinoma Using Bioinformatics Strategy: A Study Based on TCGA and GEO Datasets.Biomed Res Int. 2019 Aug 7;2019:7376034. doi: 10.1155/2019/7376034. eCollection 2019. Biomed Res Int. 2019. PMID: 31485443 Free PMC article.
-
MYCT1 in cancer development: Gene structure, regulation, and biological implications for diagnosis and treatment.Biomed Pharmacother. 2023 Sep;165:115208. doi: 10.1016/j.biopha.2023.115208. Epub 2023 Jul 25. Biomed Pharmacother. 2023. PMID: 37499454 Review.
Cited by
-
The pro-invasive factor COL6A2 serves as a novel prognostic marker of glioma.Front Oncol. 2022 Nov 25;12:897042. doi: 10.3389/fonc.2022.897042. eCollection 2022. Front Oncol. 2022. PMID: 36505882 Free PMC article.
-
Clinical Impact of C-myc Oncogenic Diversity on Solid and Lymphoid Malignancies.Maedica (Bucur). 2024 Jun;19(2):355-359. doi: 10.26574/maedica.2024.19.2.355. Maedica (Bucur). 2024. PMID: 39188831 Free PMC article. Review.
-
A new risk model for CSTA, FAM83A, and MYCT1 predicts poor prognosis and is related to immune infiltration in lung squamous cell carcinoma.Am J Transl Res. 2022 Nov 15;14(11):7705-7725. eCollection 2022. Am J Transl Res. 2022. PMID: 36505278 Free PMC article.
-
Identification of Genetic Risk Factors for Keratinocyte Cancer in Immunosuppressed Solid Organ Transplant Recipients: A Case-Control Study.Cancers (Basel). 2023 Jun 26;15(13):3354. doi: 10.3390/cancers15133354. Cancers (Basel). 2023. PMID: 37444464 Free PMC article.
-
Sevoflurane promotes the apoptosis of laryngeal squamous cell carcinoma in-vitro and inhibits its malignant progression via miR-26a/FOXO1 axis.Bioengineered. 2021 Dec;12(1):6364-6376. doi: 10.1080/21655979.2021.1962684. Bioengineered. 2021. PMID: 34511023 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

