Development and Validation of a Personalized Survival Prediction Model for Uterine Adenosarcoma: A Population-Based Deep Learning Study
- PMID: 33680946
- PMCID: PMC7930479
- DOI: 10.3389/fonc.2020.623818
Development and Validation of a Personalized Survival Prediction Model for Uterine Adenosarcoma: A Population-Based Deep Learning Study
Abstract
Background: The aim was to develop a personalized survival prediction deep learning model for adenosarcoma patients using the surveillance, epidemiology and end results (SEER) database.
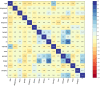
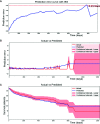
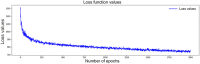
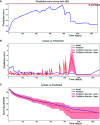
Methods: A total of 797 uterine adenosarcoma patients were enrolled in this study. Duplicated and useless variables were excluded, and 15 variables were selected for further analyses, including age, grade, positive lymph nodes or not, marital status, race, tumor extension, stage, and surgery or not. We created our deep survival learning (DSL) model to manipulate the data, which was randomly split into a training set (n = 519, 65%), validation set (n = 143, 18%) and testing set (n = 143, 18%). The Cox proportional hazard (CPH) model was also included comparatively. Finally, personalized survival curves were plotted for randomly selected patients.
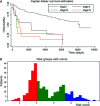
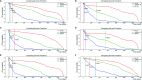
Results: The c-index for the CPH model was 0.726, and the Brier score was 0.17. For our deep survival learning model, we achieved a c-index of 0.774 and a Brier score of 0.14 in the external testing set. In addition, the limitations of the traditional staging system were revealed, and a personalized survival prediction system based on our risk scoring grouping was developed.
Conclusions: Our study developed a deep neural network model for adenosarcoma. The performance of this model was superior to that of the traditional Cox proportional hazard model. In addition, a personalized survival prediction system was developed based on our deep survival learning model, which provided more accurate prognostic information for adenosarcoma patients.
Keywords: adenosarcoma; artificial intelligence; database; deep learning; personalized model; survival prediction.
Copyright © 2021 Qu, Liu, Jiao, Zhang, Wang, Li, Dong and Cui.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Development and validation of a deep learning survival model for cervical adenocarcinoma patients.BMC Bioinformatics. 2023 Apr 13;24(1):146. doi: 10.1186/s12859-023-05239-7. BMC Bioinformatics. 2023. PMID: 37055729 Free PMC article.
-
Development and validation of survival prediction model for gastric adenocarcinoma patients using deep learning: A SEER-based study.Front Oncol. 2023 Mar 7;13:1131859. doi: 10.3389/fonc.2023.1131859. eCollection 2023. Front Oncol. 2023. PMID: 36959782 Free PMC article.
-
A novel deep learning prognostic system improves survival predictions for stage III non-small cell lung cancer.Cancer Med. 2022 Nov;11(22):4246-4255. doi: 10.1002/cam4.4782. Epub 2022 May 2. Cancer Med. 2022. PMID: 35491970 Free PMC article.
-
Comparison of deep learning models to traditional Cox regression in predicting survival of colon cancer: Based on the SEER database.J Gastroenterol Hepatol. 2024 Sep;39(9):1816-1826. doi: 10.1111/jgh.16598. Epub 2024 May 9. J Gastroenterol Hepatol. 2024. PMID: 38725241
-
Development of a Novel Deep Learning-Based Prediction Model for the Prognosis of Operable Cervical Cancer.Comput Math Methods Med. 2022 Nov 26;2022:4364663. doi: 10.1155/2022/4364663. eCollection 2022. Comput Math Methods Med. 2022. PMID: 36471752 Free PMC article.
Cited by
-
Case Report: Uterine Adenosarcoma With Sarcomatous Overgrowth and Malignant Heterologous Elements.Front Med (Lausanne). 2022 Jan 10;8:819141. doi: 10.3389/fmed.2021.819141. eCollection 2021. Front Med (Lausanne). 2022. PMID: 35083260 Free PMC article.
-
Development and validation of a deep learning survival model for cervical adenocarcinoma patients.BMC Bioinformatics. 2023 Apr 13;24(1):146. doi: 10.1186/s12859-023-05239-7. BMC Bioinformatics. 2023. PMID: 37055729 Free PMC article.
-
A novel risk classification system based on the eighth edition of TNM frameworks for esophageal adenocarcinoma patients: A deep learning approach.Front Oncol. 2022 Dec 7;12:887841. doi: 10.3389/fonc.2022.887841. eCollection 2022. Front Oncol. 2022. PMID: 36568200 Free PMC article.
-
Next-Generation Sequencing Analysis of 3 Uterine Adenosarcomas with Heterogeneously Differentiated Genomic Mutations.Int J Anal Chem. 2023 Sep 28;2023:7436368. doi: 10.1155/2023/7436368. eCollection 2023. Int J Anal Chem. 2023. PMID: 37810911 Free PMC article.
-
A deep-learning-based clinical risk stratification for overall survival in adolescent and young adult women with breast cancer.J Cancer Res Clin Oncol. 2023 Sep;149(12):10423-10433. doi: 10.1007/s00432-023-04955-0. Epub 2023 Jun 5. J Cancer Res Clin Oncol. 2023. PMID: 37277578 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

