Listeriosis Cases and Genetic Diversity of Their L. monocytogenes Isolates in China, 2008-2019
- PMID: 33680989
- PMCID: PMC7933659
- DOI: 10.3389/fcimb.2021.608352
Listeriosis Cases and Genetic Diversity of Their L. monocytogenes Isolates in China, 2008-2019
Abstract
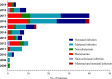
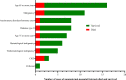
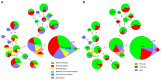
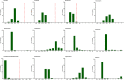
Listeriosis, caused by Listeria monocytogenes, is a severe food-borne infection. The nationwide surveillance in China concerning listeriosis is urgently needed. In the present study, 144 L. monocytogenes isolates were collected from the samples of blood, cerebrospinal fluid (CSF), and fetal membrane/placenta in China for 12 years from 2008 to 2019. We summarized these listeriosis patients' demographical and clinical features and outcomes. The susceptibility profile for 12 antibiotics was also determined by the broth microdilution method. Multilocus sequence typing (MLST) and serogroups of these listeria isolates were analyzed to designate epidemiological types. We enrolled 144 cases from 29 healthcare centers, including 96 maternal-neonatal infections, 33 cases of bacteremia, 13 cases of neurolisteriosis, and two cutaneous listeriosis. There were 31 (59.6%) fetal loss in 52 pregnant women and four (9.8%) neonatal death in 41 newborns. Among the 48 nonmaternal-neonatal cases, 12.5% (6/48) died, 41.7% (20/48) were female, and 64.6% (31/48) occurred in those with significant comorbidities. By MLST, the strains were distinguished into 23 individual sequence types (STs). The most prevalent ST was ST87 (49 isolates, 34.0%), followed by ST1 (18, 12.5%), ST8 (10, 6.9%), ST619 (9, 6.3%), ST7 (7, 4.9%) and ST3 (7, 4.9%). Furthermore, all L. monocytogenes isolates were uniformly susceptible to penicillin, ampicillin, and meropenem. In summary, our study highlights a high genotypic diversity of L. monocytogenes strains causing clinical listeriosis in China. Furthermore, a high prevalence of ST87 and ST1 in the listeriosis should be noted.
Keywords: Listeria monocytogenes; antibiotic resistance profile; isteriosis; multilocus sequence typing; neonatal listeriosis.
Copyright © 2021 Lu, Yang, Gao, Li, Cui, Huang, Chen, Wang, Wang, Liu, Li, Zhang, Jiao, Xu, Song, Fu, Xu, Yang, Ning, Wang, Bao, Luo, Wu, Yang, Li, Tang, Wang, Guo, Zeng and Zhong.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Genetic diversity and molecular typing of Listeria monocytogenes in China.BMC Microbiol. 2012 Jun 22;12:119. doi: 10.1186/1471-2180-12-119. BMC Microbiol. 2012. PMID: 22727037 Free PMC article.
-
Prevalence, Genotypic Characteristics, and Antibiotic Resistance of Listeria monocytogenes From Retail Foods in Huzhou, China.J Food Prot. 2024 Jul;87(7):100307. doi: 10.1016/j.jfp.2024.100307. Epub 2024 May 24. J Food Prot. 2024. PMID: 38797247
-
Genetic diversity, virulence factors, and antimicrobial resistance of Listeria monocytogenes from food, livestock, and clinical samples between 2002 and 2019 in China.Int J Food Microbiol. 2022 Apr 2;366:109572. doi: 10.1016/j.ijfoodmicro.2022.109572. Epub 2022 Feb 7. Int J Food Microbiol. 2022. PMID: 35176609
-
The Epidemiology of Listeria monocytogenes in China.Foodborne Pathog Dis. 2018 Aug;15(8):459-466. doi: 10.1089/fpd.2017.2409. Foodborne Pathog Dis. 2018. PMID: 30124341 Review.
-
Human Listeriosis.Clin Microbiol Rev. 2023 Mar 23;36(1):e0006019. doi: 10.1128/cmr.00060-19. Epub 2022 Dec 8. Clin Microbiol Rev. 2023. PMID: 36475874 Free PMC article. Review.
Cited by
-
Analysis of Clinical and Microbiological Features of Listeria monocytogenes Infection.Infect Drug Resist. 2023 May 8;16:2793-2803. doi: 10.2147/IDR.S408089. eCollection 2023. Infect Drug Resist. 2023. PMID: 37187483 Free PMC article.
-
DegU-mediated suppression of carbohydrate uptake in Listeria monocytogenes increases adaptation to oxidative stress.Appl Environ Microbiol. 2023 Oct 31;89(10):e0101723. doi: 10.1128/aem.01017-23. Epub 2023 Oct 3. Appl Environ Microbiol. 2023. PMID: 37787570 Free PMC article.
-
Genomic Characterization of a Clinical Listeria monocytogenes ST1 Isolate Recovered from the Blood Sample of a Woman with Third Trimester Stillbirth.Infect Drug Resist. 2022 Sep 19;15:5529-5532. doi: 10.2147/IDR.S384589. eCollection 2022. Infect Drug Resist. 2022. PMID: 36158236 Free PMC article.
-
Dissecting Listeria monocytogenes Persistent Contamination in a Retail Market Using Whole-Genome Sequencing.Microbiol Spectr. 2022 Jun 29;10(3):e0018522. doi: 10.1128/spectrum.00185-22. Epub 2022 May 17. Microbiol Spectr. 2022. PMID: 35579473 Free PMC article.
-
An Update Review on Listeria Infection in Pregnancy.Infect Drug Resist. 2021 May 26;14:1967-1978. doi: 10.2147/IDR.S313675. eCollection 2021. Infect Drug Resist. 2021. PMID: 34079306 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

