Comparison of the Inhibitory Binding Modes Between the Planar Fascaplysin and Its Nonplanar Tetrahydro-β-carboline Analogs in CDK4
- PMID: 33681142
- PMCID: PMC7930575
- DOI: 10.3389/fchem.2021.614154
Comparison of the Inhibitory Binding Modes Between the Planar Fascaplysin and Its Nonplanar Tetrahydro-β-carboline Analogs in CDK4
Abstract
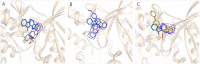
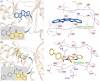
Fascaplysin is a natural marine product originating from sponges, attracting widespread attention due to its potential inhibitory activities against CDK4. However, its clinical application has been largely limited because of serious adverse effects caused by planar skeleton. To reduce the serious adverse effects, 18 tetrahydro-β-carboline analogs (compounds 6a-i and 7a-i) were designed and synthesized via breaking the planarity of fascaplysin, and the biological activities of the synthesized compounds were evaluated by MTT assay and CDK4/CycD3 enzyme inhibition assay. The title compounds showed varying degrees of inhibitory activities, especially the cytotoxicity of compound 6c against HeLa cells (IC50 = 1.03 ± 0.19 μM) with quite weak cytotoxicity toward the normal cells WI-38 (IC50 = 311.51 ± 56.06 μM), and the kinase inhibition test indicated that compound 6c was a potential CDK4 inhibitor. In order to further compare the action mechanisms of planar and nonplanar molecules on CDK4, the studied complexes of CDK4 bound with fascaplysin and three representative compounds (compound 6a-c) with bioactivities gradient were constructed by molecular docking and further verified through molecular dynamic simulation, which identified the key residues contributing to the ligands' binding. By comparing the binding modes of the constructed systems, it could be found that the residues contributing significantly to compound 6c's binding were highly consistent with those contributing significantly to fascaplysin's binding. Through the design, synthesis of the nonplanar fascaplysin derivatives, and binding mechanism analysis, some valuable hints for the discovery of antitumor drug candidates could be provided.
Keywords: MD simulation; MTT assay; fascaplysin; molecular docking; nonplanar.
Copyright © 2021 Liang, Quan, Bu, Li, Liu, Wang, He, Jia and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
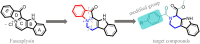





Similar articles
-
Marine-Derived Lead Fascaplysin: Pharmacological Activity, Total Synthesis, and Structural Modification.Mar Drugs. 2023 Mar 31;21(4):226. doi: 10.3390/md21040226. Mar Drugs. 2023. PMID: 37103365 Free PMC article. Review.
-
Discovery of a marine-derived bis-indole alkaloid fascaplysin, as a new class of potent P-glycoprotein inducer and establishment of its structure-activity relationship.Eur J Med Chem. 2016 Jan 1;107:1-11. doi: 10.1016/j.ejmech.2015.10.049. Epub 2015 Oct 31. Eur J Med Chem. 2016. PMID: 26560048
-
Synthesis, crystal structure and biological activity of beta-carboline based selective CDK4-cyclin D1 inhibitors.Org Biomol Chem. 2006 Dec 21;4(24):4478-84. doi: 10.1039/b613861f. Epub 2006 Nov 9. Org Biomol Chem. 2006. PMID: 17268643
-
Inhibition of cyclin-dependent kinase 4 (Cdk4) by fascaplysin, a marine natural product.Biochem Biophys Res Commun. 2000 Sep 7;275(3):877-84. doi: 10.1006/bbrc.2000.3349. Biochem Biophys Res Commun. 2000. PMID: 10973815
-
Chemistry and biology of fascaplysin, a potent marine-derived CDK-4 inhibitor.Mini Rev Med Chem. 2012 Jun;12(7):650-64. doi: 10.2174/138955712800626719. Mini Rev Med Chem. 2012. PMID: 22512549 Review.
Cited by
-
Mechanisms Underlying Gastrodin Alleviating Vincristine-Induced Peripheral Neuropathic Pain.Front Pharmacol. 2021 Dec 16;12:744663. doi: 10.3389/fphar.2021.744663. eCollection 2021. Front Pharmacol. 2021. PMID: 34975470 Free PMC article.
-
Marine-Derived Lead Fascaplysin: Pharmacological Activity, Total Synthesis, and Structural Modification.Mar Drugs. 2023 Mar 31;21(4):226. doi: 10.3390/md21040226. Mar Drugs. 2023. PMID: 37103365 Free PMC article. Review.
References
-
- AMBER v. 16 (2016). San Francisco, CA: University of California.
LinkOut - more resources
Full Text Sources
Other Literature Sources

