PCR Array Technology in Biopsy Samples Identifies Up-Regulated mTOR Pathway Genes as Potential Rejection Biomarkers After Kidney Transplantation
- PMID: 33681239
- PMCID: PMC7927668
- DOI: 10.3389/fmed.2021.547849
PCR Array Technology in Biopsy Samples Identifies Up-Regulated mTOR Pathway Genes as Potential Rejection Biomarkers After Kidney Transplantation
Abstract
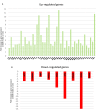
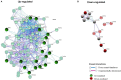
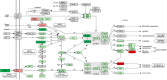
Background: Antibody-mediated rejection (AMR) is the major cause of kidney transplant rejection. The donor-specific human leukocyte antigen (HLA) antibody (DSA) response to a renal allograft is not fully understood yet. mTOR complex has been described in the accommodation or rejection of transplants and integrates responses from a wide variety of signals. The aim of this study was to analyze the expression of the mTOR pathway genes in a large cohort of kidney transplant patients to determine its possible influence on the transplant outcome. Methods: A total of 269 kidney transplant patients monitored for DSA were studied. The patients were divided into two groups, one with recipients that had transplant rejection (+DSA/+AMR) and a second group of recipients without rejection (+DSA/-AMR and -DSA/-AMR, controls). Total RNA was extracted from kidney biopsies and reverse transcribed to cDNA. Human mTOR-PCR array technology was used to determine the expression of 84 mTOR pathway genes. STRING and REVIGO software were used to simulate gene to gene interaction and to assign a molecular function. Results: The studied groups showed a different expression of the mTOR pathway related genes. Recipients that had transplant rejection showed an over-expressed transcript (≥5-fold) of AKT1S1, DDIT4, EIF4E, HRAS, IGF1, INS, IRS1, PIK3CD, PIK3CG, PRKAG3, PRKCB (>12-fold), PRKCG, RPS6KA2, TELO2, ULK1, and VEGFC, compared with patients that did not have rejection. AKT1S1 transcripts were more expressed in +DSA/-AMR biopsies compared with +DSA/+AMR. The main molecular functions of up-regulated gene products were phosphotransferase activity, insulin-like grown factor receptor and ribonucleoside phosphate binding. The group of patients with transplant rejection also showed an under-expressed transcript (≥5-fold) of VEGFA (>15-fold), RPS6, and RHOA compared with the group without rejection. The molecular function of down-regulated gene products such as protein kinase activity and carbohydrate derivative binding proteins was also analyzed. Conclusions: We have found a higher number of over-expressed mTOR pathway genes than under-expressed ones in biopsies from rejected kidney transplants (+DSA/+AMR) with respect to controls. In addition to this, the molecular function of both types of transcripts (over/under expressed) is different. Therefore, further studies are needed to determine if variations in gene expression profiles can act as predictors of graft loss, and a better understanding of the mechanisms of action of the involved proteins would be necessary.
Keywords: Gene expression; PCR array; antibody-mediated rejection; mTOR; medico-legal autopsy.
Copyright © 2021 Legaz, Bernardo, Alfaro, Martínez-Banaclocha, Galián, Jimenez-Coll, Boix, Mrowiec, Salmeron, Botella, Parrado, Moya-Quiles, Minguela, Llorente, Peña-Moral and Muro.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Muro M, Llorente S, Marín L, Moya-Quiles MR, Gonzalez-Soriano MJ, Prieto A, et al. . Acute vascular rejection mediated by HLA antibodies in a cadaveric kidney recipient: discrepancies between flow PRA, ELISA and CDC vs. luminex screening. Nephrol Dial Transplant. (2005) 20:223–6. 10.1093/ndt/gfh527 - DOI - PubMed
-
- Bosch A, Llorente S, Diaz JA, Salgado G, López M, Boix F, et al. . Low median fluorescence intensity could be a nonsafety concept of immunologic risk evaluation in patients with shared molecular eplets in kidney transplantation. Hum Immunol. (2012) 73:522–5. 10.1016/j.humimm.2012.02.020 - DOI - PubMed
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

