Effect of Lauric Acid on the Stability of Aβ42 Oligomers
- PMID: 33681618
- PMCID: PMC7931375
- DOI: 10.1021/acsomega.0c06211
Effect of Lauric Acid on the Stability of Aβ42 Oligomers
Abstract
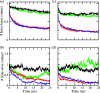
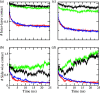
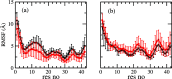
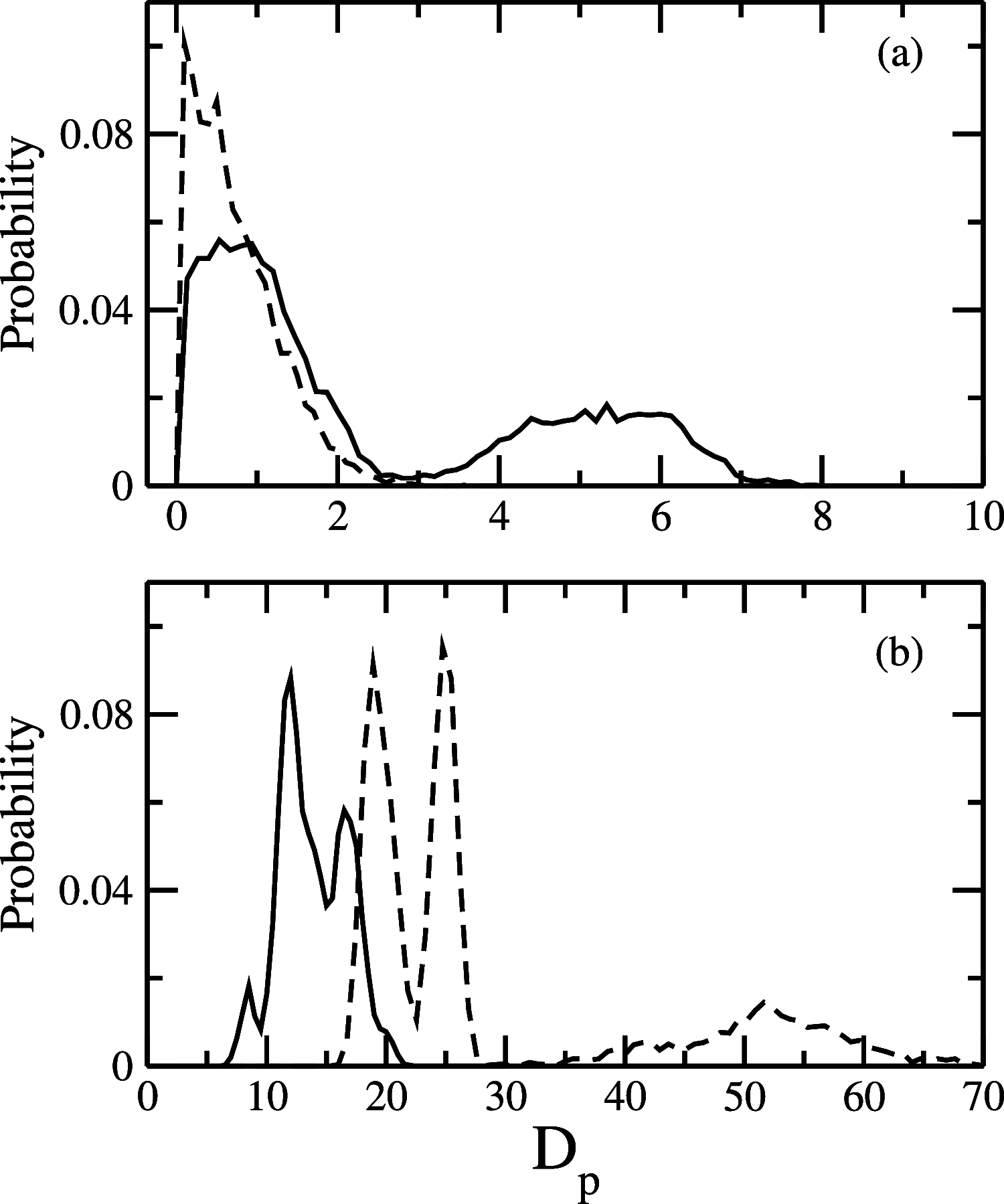
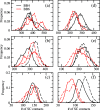
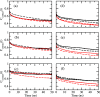
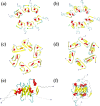
While Alzheimer's disease is correlated with the presence of Aβ fibrils in patient brains, the more likely agents are their precursors, soluble oligomers that may form pores or otherwise distort cell membranes. Using all-atom molecular dynamics simulation, we study how the presence of fatty acids such as lauric acid changes the stability of pore-forming oligomers built from three-stranded Aβ42 chains. Such a change would alter the distribution of amyloids in the fatty acid-rich brain environment and therefore could explain the lower polymorphism observed in Aβ fibrils derived from brains of patients with Alzheimer's disease. We find that lauric acid stabilizes both ring-like and barrel-shaped models, with the effect being stronger for barrel-like models than for ring-like oligomers.
© 2021 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

