The new SARS-CoV-2 strain shows a stronger binding affinity to ACE2 due to N501Y mutant
- PMID: 33681755
- PMCID: PMC7923861
- DOI: 10.1016/j.medidd.2021.100086
The new SARS-CoV-2 strain shows a stronger binding affinity to ACE2 due to N501Y mutant
Abstract
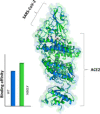
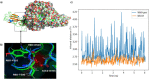
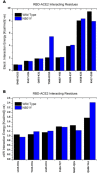
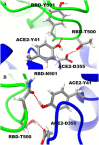
SARS-CoV-2 is a global challenge due to its ability to spread much faster than the SARS-CoV, which was attributed to the mutations in the receptor binding domain (RBD). These mutations enhanced the electrostatic interactions. Recently, a new strain is reported in the UK that includes a mutation (N501Y) in the RBD, that is possibly increasing the infection rate. Here, using Molecular Dynamics simulations (MD) and Monte Carlo (MC) sampling, we show that the N501 mutation enhanced the electrostatic interactions due to the formation of a strong hydrogen bond between SARS-CoV-2-T500 and ACE2-D355 near the mutation site. In addition, we observed that the electrostatic interactions between the SARS-CoV-2 and ACE2 in the wild type and the mutant are dominated by salt-bridges formed between SARS-CoV-2-K417 and ACE2-D30, SARS-CoV-2-K458, ACE2-E23, and SARS-CoV-2-R403 and ACE2-E37. These interactions contributed more than 40% of the total binding energies.
Keywords: Binding domains; Electrostatic interactions; Molecular dynamics; Monte Carlo; SARS-CoV; SARS-CoV-2.
© 2021 The Author(s).
Conflict of interest statement
None.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
