Contrasting transcriptional responses to Fusarium virguliforme colonization in symptomatic and asymptomatic hosts
- PMID: 33681966
- PMCID: PMC8136916
- DOI: 10.1093/plcell/koaa021
Contrasting transcriptional responses to Fusarium virguliforme colonization in symptomatic and asymptomatic hosts
Abstract
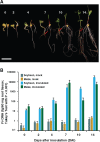
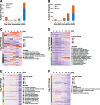
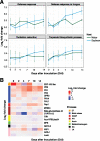
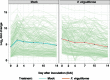
The broad host range of Fusarium virguliforme represents a unique comparative system to identify and define differentially induced responses between an asymptomatic monocot host, maize (Zea mays), and a symptomatic eudicot host, soybean (Glycine max). Using a temporal, comparative transcriptome-based approach, we observed that early gene expression profiles of root tissue from infected maize suggest that pathogen tolerance coincides with the rapid induction of senescence dampening transcriptional regulators, including ANACs (Arabidopsis thaliana NAM/ATAF/CUC protein) and Ethylene-Responsive Factors. In contrast, the expression of senescence-associated processes in soybean was coincident with the appearance of disease symptom development, suggesting pathogen-induced senescence as a key pathway driving pathogen susceptibility in soybean. Based on the analyses described herein, we posit that root senescence is a primary contributing factor underlying colonization and disease progression in symptomatic versus asymptomatic host-fungal interactions. This process also supports the lifestyle and virulence of F. virguliforme during biotrophy to necrotrophy transitions. Further support for this hypothesis lies in comprehensive co-expression and comparative transcriptome analyses, and in total, supports the emerging concept of necrotrophy-activated senescence. We propose that F. virguliforme conditions an environment within symptomatic hosts, which favors susceptibility through transcriptomic reprogramming, and as described herein, the induction of pathways associated with senescence during the necrotrophic stage of fungal development.
© The Author(s) 2020. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Figures




References
-
- Alexa A, Rahnenfuhrer J (2018) topGO: Enrichment analysis for gene ontology. R package version 2.36.0
-
- Allardyce JA, Rookes JE, Hussain HI, Cahill DM (2013) Transcriptional profiling of Zea mays roots reveals roles for jasmonic acid and terpenoids in resistance against Phytophthora cinnamomi. Funct Integr Genom 13: 217–228 - PubMed
-
- Allen TW, Bradley CA, Sisson AJ, Byamukama E, Chilvers MI, Coker CM, Collins AA, Damicone JP, Dorrance AE, Dufault NS, et al. (2017) Soybean yield loss estimates due to diseases in the United States and Ontario, Canada, from 2010 to 2014. Plant Health Prog 18: 19–27
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

