High-Throughput Quality Control Assay for the Solid-Phase Synthesis of DNA-Encoded Libraries of Macrocycles*
- PMID: 33682283
- PMCID: PMC8193836
- DOI: 10.1002/anie.202100230
High-Throughput Quality Control Assay for the Solid-Phase Synthesis of DNA-Encoded Libraries of Macrocycles*
Abstract
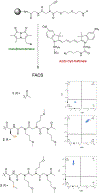
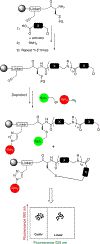
There is considerable interest in the development of libraries of scaffold-diverse macrocycles as a source of ligands for difficult targets, such as protein-protein interaction surfaces. A classic problem in the synthesis of high-quality macrocyclic libraries is that some linear precursors will cyclize efficiently while some will not, depending on their conformational preferences. We report here a powerful quality control method that can be employed to readily distinguish between scaffolds that do and do not cyclize efficiently during solid-phase synthesis of thioether macrocycles without the need for tedious liquid chromatography/mass spectrometry analysis. We demonstrate that this assay can be employed to identify linear impurities in a DNA-encoded library of macrocycles. We also use the method to establish a useful quality control protocol for re-synthesis of putative macrocyclic screening hits.
Keywords: DNA-encoded libraries; macrocycles; peptidomimetics.
© 2021 Wiley-VCH GmbH.
Conflict of interest statement
Conflict of interest
T.K. is a major shareholder in Deluge Biotechnologies, which is commercializing DNA-encoded libraries of macrocycles.
Figures




References
-
- Driggers EM, Hale SP, Lee J, Terrett NK, Nat. Rev. Drug Discovery 2008, 7, 608–624; - PubMed
- Whitty A, Viarengo LA, Zhong M, Org. Biomol. Chem 2017, 15, 7729–7735; - PubMed
- Bauer RA, Wurst JM, Tan DS, Curr. Opin. Chem. Biol 2010, 14, 308–314; - PMC - PubMed
- Dandapani S, Marcaurelle LA, Nat. Chem. Biol 2010, 6, 861–863. - PubMed
-
- Villar EA, Beglov D, Chennamadhavuni S, Porco JA, Kozakov D, Vajda S, Whitty A, Nat. Chem. Biol 2014, 10, 723–731; - PMC - PubMed
- Over B, Matsson P, Tyrchan C, Artursson P, Doak BC, Foley MA, Hilgendorf C, Johnston SE, Lee MD, Lewis RJ, McCarren P, Muncipinto G, Norinder U, Perry MWD, Duvall JR, Kihlberg J, Nat. Chem. Biol 2016, 12, 1065–1074; - PubMed
- Pye CR, Hewitt WM, Schwochert J, Haddad TD, Townsend CE, Etienne L, Lao Y, Limberakis C, Furukawa A, Mathiowetz AM, Price DA, Liras S, Lokey RS, J. Med. Chem 2017, 60, 1665–1672. - PMC - PubMed
-
- Connors WH, Hale SP, Terrett NK, Curr. Opin. Chem. Biol 2015, 26, 42–47. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

