Epithelial miR-141 regulates IL-13-induced airway mucus production
- PMID: 33682796
- PMCID: PMC8021117
- DOI: 10.1172/jci.insight.139019
Epithelial miR-141 regulates IL-13-induced airway mucus production
Abstract
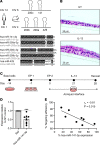
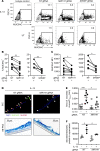
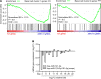
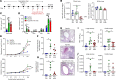
IL-13-induced goblet cell metaplasia contributes to airway remodeling and pathological mucus hypersecretion in asthma. miRNAs are potent modulators of cellular responses, but their role in mucus regulation is largely unexplored. We hypothesized that airway epithelial miRNAs play roles in IL-13-induced mucus regulation. miR-141 is highly expressed in human and mouse airway epithelium, is altered in bronchial brushings from asthmatic subjects at baseline, and is induced shortly after airway allergen exposure. We established a CRISPR/Cas9-based protocol to target miR-141 in primary human bronchial epithelial cells that were differentiated at air-liquid-interface, and goblet cell hyperplasia was induced by IL-13 stimulation. miR-141 disruption resulted in decreased goblet cell frequency, intracellular MUC5AC, and total secreted mucus. These effects correlated with a reduction in a goblet cell gene expression signature and enrichment of a basal cell gene expression signature defined by single cell RNA sequencing. Furthermore, intranasal administration of a sequence-specific mmu-miR-141-3p inhibitor in mice decreased Aspergillus-induced secreted mucus and mucus-producing cells in the lung and reduced airway hyperresponsiveness without affecting cellular inflammation. In conclusion, we have identified a miRNA that regulates pathological airway mucus production and is amenable to therapeutic manipulation through an inhaled route.
Keywords: Asthma; Noncoding RNAs; Pulmonology; Th2 response.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

