Aggressiveness and Metastatic Potential of Breast Cancer Cells Co-Cultured with Preadipocytes and Exposed to an Environmental Pollutant Dioxin: An in Vitro and in Vivo Zebrafish Study
- PMID: 33683140
- PMCID: PMC7939125
- DOI: 10.1289/EHP7102
Aggressiveness and Metastatic Potential of Breast Cancer Cells Co-Cultured with Preadipocytes and Exposed to an Environmental Pollutant Dioxin: An in Vitro and in Vivo Zebrafish Study
Abstract
Background: Breast cancer (BC) is a major public health concern, and its prognosis is very poor once metastasis occurs. The tumor microenvironment and chemical pollution have been suggested recently to contribute, independently, to the development of metastatic cells. The BC microenvironment consists, in part, of adipocytes and preadipocytes in which persistent organic pollutants (POPs) can be stored.
Objectives: We aimed to test the hypothesis that these two factors (2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), an extensively studied, toxic POP and the microenvironment) may interact to increase tumor aggressiveness.
Methods: We used a co-culture model using BC MCF-7 cells or MDA-MB-231 cells together with hMADS preadipocytes to investigate the contribution of the microenvironment and 2,3,7,8-tetrachlorodibenzo-p-dioxin TCDD on BC cells. Global differences were characterized using a high-throughput proteomic assay. Subsequently we measured the BC stem cell-like activity, analyzed the cell morphology, and used a zebrafish larvae model to study the metastatic potential of the BC cells.
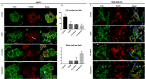
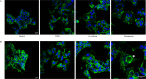
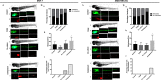
Results: We found that coexposure to TCDD and preadipocytes modified BC cell properties; moreover, it induced the expression of ALDH1A3, a cancer stem cell marker, and the appearance of giant cancer cells with cell-in-cell structures (CICs), which are associated with malignant metastatic progression, that we demonstrated in vivo.
Discussion: The results of our study using BC cell lines co-cultured with preadipocytes and a POP and an in vivo zebrafish model of metastasis suggest that the interactions between BC cells and their microenvironment could affect their invasive or metastatic potential. https://doi.org/10.1289/EHP7102.
Figures



References
-
- Abdalla F, Jamela B, Rabia M, Hussein H, Abdelbaset B, Yrjö C. 2009. Correlation of nuclear morphometry of breast cancer in histological sections with clinicopathological features and prognosis. Anticancer Res 29(5):1771–1776. PMID: 19443402. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

