Globetrotting strangles: the unbridled national and international transmission of Streptococcus equi between horses
- PMID: 33684029
- PMCID: PMC8190609
- DOI: 10.1099/mgen.0.000528
Globetrotting strangles: the unbridled national and international transmission of Streptococcus equi between horses
Abstract
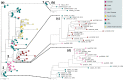
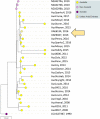
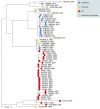
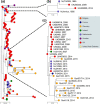
The equine disease strangles, which is characterized by the formation of abscesses in the lymph nodes of the head and neck, is one of the most frequently diagnosed infectious diseases of horses around the world. The causal agent, Streptococcus equi subspecies equi, establishes a persistent infection in approximately 10 % of animals that recover from the acute disease. Such 'carrier' animals appear healthy and are rarely identified during routine veterinary examinations pre-purchase or transit, but can transmit S. equi to naïve animals initiating new episodes of disease. Here, we report the analysis and visualization of phylogenomic and epidemiological data for 670 isolates of S. equi recovered from 19 different countries using a new core-genome multilocus sequence typing (cgMLST) web bioresource. Genetic relationships among all 670 S. equi isolates were determined at high resolution, revealing national and international transmission events that drive this endemic disease in horse populations throughout the world. Our data argue for the recognition of the international importance of strangles by the Office International des Épizooties to highlight the health, welfare and economic cost of this disease. The Pathogenwatch cgMLST web bioresource described herein is available for tailored genomic analysis of populations of S. equi and its close relative S. equi subspecies zooepidemicus that are recovered from horses and other animals, including humans, throughout the world. This article contains data hosted by Microreact.
Keywords: Streptococcus equi; genome diversity; pandemic; strangles; transmission.
Conflict of interest statement
A.S.W. is employed by Intervacc AB.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

