A computational study of co-inhibitory immune complex assembly at the interface between T cells and antigen presenting cells
- PMID: 33684103
- PMCID: PMC7971848
- DOI: 10.1371/journal.pcbi.1008825
A computational study of co-inhibitory immune complex assembly at the interface between T cells and antigen presenting cells
Abstract
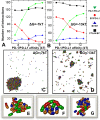
The activation and differentiation of T-cells are mainly directly by their co-regulatory receptors. T lymphocyte-associated protein-4 (CTLA-4) and programed cell death-1 (PD-1) are two of the most important co-regulatory receptors. Binding of PD-1 and CTLA-4 with their corresponding ligands programed cell death-ligand 1 (PD-L1) and B7 on the antigen presenting cells (APC) activates two central co-inhibitory signaling pathways to suppress T cell functions. Interestingly, recent experiments have identified a new cis-interaction between PD-L1 and B7, suggesting that a crosstalk exists between two co-inhibitory receptors and the two pairs of ligand-receptor complexes can undergo dynamic oligomerization. Inspired by these experimental evidences, we developed a coarse-grained model to characterize the assembling of an immune complex consisting of CLTA-4, B7, PD-L1 and PD-1. These four proteins and their interactions form a small network motif. The temporal dynamics and spatial pattern formation of this network was simulated by a diffusion-reaction algorithm. Our simulation method incorporates the membrane confinement of cell surface proteins and geometric arrangement of different binding interfaces between these proteins. A wide range of binding constants was tested for the interactions involved in the network. Interestingly, we show that the CTLA-4/B7 ligand-receptor complexes can first form linear oligomers, while these oligomers further align together into two-dimensional clusters. Similar phenomenon has also been observed in other systems of cell surface proteins. Our test results further indicate that both co-inhibitory signaling pathways activated by B7 and PD-L1 can be down-regulated by the new cis-interaction between these two ligands, consistent with previous experimental evidences. Finally, the simulations also suggest that the dynamic and the spatial properties of the immune complex assembly are highly determined by the energetics of molecular interactions in the network. Our study, therefore, brings new insights to the co-regulatory mechanisms of T cell activation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

