Genome-Wide fitness analysis of group B Streptococcus in human amniotic fluid reveals a transcription factor that controls multiple virulence traits
- PMID: 33684178
- PMCID: PMC7971860
- DOI: 10.1371/journal.ppat.1009116
Genome-Wide fitness analysis of group B Streptococcus in human amniotic fluid reveals a transcription factor that controls multiple virulence traits
Abstract
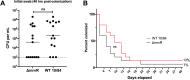
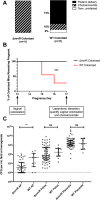
Streptococcus agalactiae (group B Streptococcus; GBS) remains a dominant cause of serious neonatal infections. One aspect of GBS that renders it particularly virulent during the perinatal period is its ability to invade the chorioamniotic membranes and persist in amniotic fluid, which is nutritionally deplete and rich in fetal immunologic factors such as antimicrobial peptides. We used next-generation sequencing of transposon-genome junctions (Tn-seq) to identify five GBS genes that promote survival in the presence of human amniotic fluid. We confirmed our Tn-seq findings using a novel CRISPR inhibition (CRISPRi) gene expression knockdown system. This analysis showed that one gene, which encodes a GntR-class transcription factor that we named MrvR, conferred a significant fitness benefit to GBS in amniotic fluid. We generated an isogenic targeted deletion of the mrvR gene, which had a growth defect in amniotic fluid relative to the wild type parent strain. The mrvR deletion strain also showed a significant biofilm defect in vitro. Subsequent in vivo studies showed that while the mutant was able to cause persistent murine vaginal colonization, pregnant mice colonized with the mrvR deletion strain did not develop preterm labor despite consistent GBS invasion of the uterus and the fetoplacental units. In contrast, pregnant mice colonized with wild type GBS consistently deliver prematurely. In a sepsis model the mrvR deletion strain showed significantly decreased lethality. In order to better understand the mechanism by which this newly identified transcription factor controls GBS virulence, we performed RNA-seq on wild type and mrvR deletion GBS strains, which revealed that the transcription factor affects expression of a wide range of genes across the GBS chromosome. Nucleotide biosynthesis and salvage pathways were highly represented among the set of differentially expressed genes, suggesting that MrvR may be involved in regulating nucleotide availability.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
The Streptococcus agalactiae Stringent Response Enhances Virulence and Persistence in Human Blood.Infect Immun. 2017 Dec 19;86(1):e00612-17. doi: 10.1128/IAI.00612-17. Print 2018 Jan. Infect Immun. 2017. PMID: 29109175 Free PMC article.
-
The Streptococcus agalactiae LytSR two-component regulatory system promotes vaginal colonization and virulence in vivo.Microbiol Spectr. 2024 Nov 5;12(11):e0197024. doi: 10.1128/spectrum.01970-24. Epub 2024 Oct 14. Microbiol Spectr. 2024. PMID: 39400158 Free PMC article.
-
Analysis of two-component systems in group B Streptococcus shows that RgfAC and the novel FspSR modulate virulence and bacterial fitness.mBio. 2014 May 20;5(3):e00870-14. doi: 10.1128/mBio.00870-14. mBio. 2014. PMID: 24846378 Free PMC article.
-
Acid Stress Response Mechanisms of Group B Streptococci.Front Cell Infect Microbiol. 2017 Sep 7;7:395. doi: 10.3389/fcimb.2017.00395. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28936424 Free PMC article. Review.
-
Biofilm formation by Streptococcus agalactiae: influence of environmental conditions and implicated virulence factors.Front Cell Infect Microbiol. 2015 Feb 4;5:6. doi: 10.3389/fcimb.2015.00006. eCollection 2015. Front Cell Infect Microbiol. 2015. PMID: 25699242 Free PMC article. Review.
Cited by
-
Gestational diabetes augments group B Streptococcus infection by disrupting maternal immunity and the vaginal microbiota.Nat Commun. 2024 Feb 3;15(1):1035. doi: 10.1038/s41467-024-45336-6. Nat Commun. 2024. PMID: 38310089 Free PMC article.
-
Coordinated regulation of osmotic imbalance by c-di-AMP shapes ß-lactam tolerance in Group B Streptococcus.Microlife. 2024 Jun 12;5:uqae014. doi: 10.1093/femsml/uqae014. eCollection 2024. Microlife. 2024. PMID: 38993744 Free PMC article.
-
Functional Genomics Identified Novel Genes Involved in Growth at Low Temperatures in Listeria monocytogenes.Microbiol Spectr. 2022 Aug 31;10(4):e0071022. doi: 10.1128/spectrum.00710-22. Epub 2022 Jun 23. Microbiol Spectr. 2022. PMID: 35735974 Free PMC article.
-
CRISPR-Based Approaches for Gene Regulation in Non-Model Bacteria.Front Genome Ed. 2022 Jun 23;4:892304. doi: 10.3389/fgeed.2022.892304. eCollection 2022. Front Genome Ed. 2022. PMID: 35813973 Free PMC article. Review.
-
Bacterial and Host Determinants of Group B Streptococcal Vaginal Colonization and Ascending Infection in Pregnancy.Front Cell Infect Microbiol. 2021 Sep 3;11:720789. doi: 10.3389/fcimb.2021.720789. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34540718 Free PMC article. Review.
References
-
- Regan JA, Kelbanoff MA, Nugent RP. The Epidemiology of Group B Streptococcal Colonization in Pregnancy. Obstetrics & Gynecology. 1991;77: 604. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

