RHO to the DOCK for GDP disembarking: Structural insights into the DOCK GTPase nucleotide exchange factors
- PMID: 33684443
- PMCID: PMC8063744
- DOI: 10.1016/j.jbc.2021.100521
RHO to the DOCK for GDP disembarking: Structural insights into the DOCK GTPase nucleotide exchange factors
Abstract
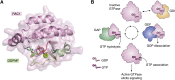
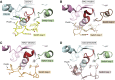
The human dedicator of cytokinesis (DOCK) family consists of 11 structurally conserved proteins that serve as atypical RHO guanine nucleotide exchange factors (RHO GEFs). These regulatory proteins act as mediators in numerous cellular cascades that promote cytoskeletal remodeling, playing roles in various crucial processes such as differentiation, migration, polarization, and axon growth in neurons. At the molecular level, DOCK DHR2 domains facilitate nucleotide dissociation from small GTPases, a process that is otherwise too slow for rapid spatiotemporal control of cellular signaling. Here, we provide an overview of the biological and structural characteristics for the various DOCK proteins and describe how they differ from other RHO GEFs and between DOCK subfamilies. The expression of the family varies depending on cell or tissue type, and they are consequently implicated in a broad range of disease phenotypes, particularly in the brain. A growing body of available structural information reveals the mechanism by which the catalytic DHR2 domain elicits nucleotide dissociation and also indicates strategies for the discovery and design of high-affinity small-molecule inhibitors. Such compounds could serve as chemical probes to interrogate the cellular function and provide starting points for drug discovery of this important class of enzymes.
Keywords: Ras homologous (RHO) small GTPases; cell signaling; dedicator of cytokinesis (DOCK); drug discovery; guanine nucleotide exchange factor; guanosine triphosphate (GTP); structural biology.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures
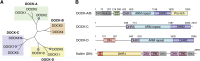




Similar articles
-
Structural biology of DOCK-family guanine nucleotide exchange factors.FEBS Lett. 2023 Mar;597(6):794-810. doi: 10.1002/1873-3468.14523. Epub 2022 Nov 4. FEBS Lett. 2023. PMID: 36271211 Free PMC article. Review.
-
Multiple factors confer specific Cdc42 and Rac protein activation by dedicator of cytokinesis (DOCK) nucleotide exchange factors.J Biol Chem. 2011 Jul 15;286(28):25341-51. doi: 10.1074/jbc.M111.236455. Epub 2011 May 24. J Biol Chem. 2011. PMID: 21613211 Free PMC article.
-
Substrate induced dynamical remodeling of the binding pocket generates GTPase specificity in DOCK family of guanine nucleotide exchange factors.Biochem Biophys Res Commun. 2022 Nov 26;631:32-40. doi: 10.1016/j.bbrc.2022.09.059. Epub 2022 Sep 20. Biochem Biophys Res Commun. 2022. PMID: 36162327
-
Activation of Rho GTPases by DOCK exchange factors is mediated by a nucleotide sensor.Science. 2009 Sep 11;325(5946):1398-402. doi: 10.1126/science.1174468. Science. 2009. PMID: 19745154
-
Targeting the Dbl and dock-family RhoGEFs: a yeast-based assay to identify cell-active inhibitors of Rho-controlled pathways.Enzymes. 2013;33 Pt A:169-91. doi: 10.1016/B978-0-12-416749-0.00008-7. Epub 2013 Aug 8. Enzymes. 2013. PMID: 25033805 Review.
Cited by
-
New Mechanisms Underlying Oncogenesis in Dbl Family Rho Guanine Nucleotide Exchange Factors.Mol Pharmacol. 2024 Aug 16;106(3):117-128. doi: 10.1124/molpharm.124.000904. Mol Pharmacol. 2024. PMID: 38902036 Review.
-
DOCK3-Associated Neurodevelopmental Disorder-Clinical Features and Molecular Basis.Genes (Basel). 2023 Oct 14;14(10):1940. doi: 10.3390/genes14101940. Genes (Basel). 2023. PMID: 37895289 Free PMC article. Review.
-
The role of RAC1 in resistance to targeted therapies in cancer.Small GTPases. 2024 Dec;15(1):1-14. doi: 10.1080/21541248.2025.2505977. Epub 2025 May 21. Small GTPases. 2024. PMID: 40396280 Free PMC article. Review.
-
Structural biology of DOCK-family guanine nucleotide exchange factors.FEBS Lett. 2023 Mar;597(6):794-810. doi: 10.1002/1873-3468.14523. Epub 2022 Nov 4. FEBS Lett. 2023. PMID: 36271211 Free PMC article. Review.
-
A multiprotein signaling complex sustains AKT and mTOR/S6K activity necessary for the survival of cancer cells undergoing stress.bioRxiv [Preprint]. 2024 Oct 1:2023.01.03.522657. doi: 10.1101/2023.01.03.522657. bioRxiv. 2024. PMID: 36711811 Free PMC article. Preprint.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

