The exo-β-N-acetylmuramidase NamZ from Bacillus subtilis is the founding member of a family of exo-lytic peptidoglycan hexosaminidases
- PMID: 33684445
- PMCID: PMC8054146
- DOI: 10.1016/j.jbc.2021.100519
The exo-β-N-acetylmuramidase NamZ from Bacillus subtilis is the founding member of a family of exo-lytic peptidoglycan hexosaminidases
Abstract
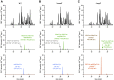
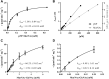
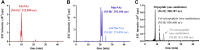
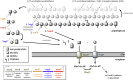
Endo-β-N-acetylmuramidases, commonly known as lysozymes, are well-characterized antimicrobial enzymes that catalyze an endo-lytic cleavage of peptidoglycan; i.e., they hydrolyze the β-1,4-glycosidic bonds connecting N-acetylmuramic acid (MurNAc) and N-acetylglucosamine (GlcNAc). In contrast, little is known about exo-β-N-acetylmuramidases, which catalyze an exo-lytic cleavage of β-1,4-MurNAc entities from the non-reducing ends of peptidoglycan chains. Such an enzyme was identified earlier in the bacterium Bacillus subtilis, but the corresponding gene has remained unknown so far. We now report that ybbC of B. subtilis, renamed namZ, encodes the reported exo-β-N-acetylmuramidase. A ΔnamZ mutant accumulated specific cell wall fragments and showed growth defects under starvation conditions, indicating a role of NamZ in cell wall turnover and recycling. Recombinant NamZ protein specifically hydrolyzed the artificial substrate para-nitrophenyl β-MurNAc and the peptidoglycan-derived disaccharide MurNAc-β-1,4-GlcNAc. Together with the exo-β-N-acetylglucosaminidase NagZ and the exo-muramoyl-l-alanine amidase AmiE, NamZ degraded intact peptidoglycan by sequential hydrolysis from the non-reducing ends. A structure model of NamZ, built on the basis of two crystal structures of putative orthologs from Bacteroides fragilis, revealed a two-domain structure including a Rossmann-fold-like domain that constitutes a unique glycosidase fold. Thus, NamZ, a member of the DUF1343 protein family of unknown function, is now classified as the founding member of a new family of glycosidases (CAZy GH171; www.cazy.org/GH171.html). NamZ-like peptidoglycan hexosaminidases are mainly present in the phylum Bacteroidetes and less frequently found in individual genomes within Firmicutes (Bacilli, Clostridia), Actinobacteria, and γ-proteobacteria.
Keywords: N-acetylglucosaminidase; N-acetylmuramidase; N-acetylmuramoyl amidase; Rossmann-fold; cell wall recycling; exo-lytic glycosidase; lysozyme; peptidoglycan hydrolase.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the content of this article.
Figures



References
-
- Warren R.A. Microbial hydrolysis of polysaccharides. Annu. Rev. Microbiol. 1996;50:183–212. - PubMed
-
- Wang J., Quirk A., Lipkowski J., Dutcher J.R., Clarke A.J. Direct in situ observation of synergism between cellulolytic enzymes during the biodegradation of crystalline cellulose fibers. Langmuir. 2013;29:14997–15005. - PubMed
-
- Itoh T., Kimoto H. Bacterial chitinase system as a model of chitin biodegradation. Adv. Exp. Med. Biol. 2019;1142:131–151. - PubMed
-
- Walter A., Mayer C. Peptidoglycan structure, biosynthesis, and dynamics during bacterial growth. In: Cohen E., Merzendorfer H., editors. Extracellular Sugar-Base Biopolymeres Matrices. Elsevier; Amsterdam, Netherlands: 2019. pp. 237–299.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

