Dihydrouridine synthesis in tRNAs is under reductive evolution in Mollicutes
- PMID: 33685366
- PMCID: PMC8632129
- DOI: 10.1080/15476286.2021.1899653
Dihydrouridine synthesis in tRNAs is under reductive evolution in Mollicutes
Abstract
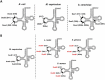
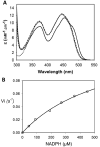
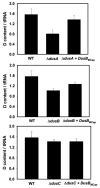
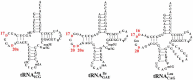
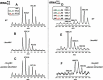
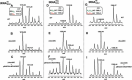
Dihydrouridine (D) is a tRNA-modified base conserved throughout all kingdoms of life and assuming an important structural role. The conserved dihydrouridine synthases (Dus) carries out D-synthesis. DusA, DusB and DusC are bacterial members, and their substrate specificity has been determined in Escherichia coli. DusA synthesizes D20/D20a while DusB and DusC are responsible for the synthesis of D17 and D16, respectively. Here, we characterize the function of the unique dus gene encoding a DusB detected in Mollicutes, which are bacteria that evolved from a common Firmicute ancestor via massive genome reduction. Using in vitro activity tests as well as in vivo E. coli complementation assays with the enzyme from Mycoplasma capricolum (DusBMCap), a model organism for the study of these parasitic bacteria, we show that, as expected for a DusB homolog, DusBMCap modifies U17 to D17 but also synthetizes D20/D20a combining therefore both E. coli DusA and DusB activities. Hence, this is the first case of a Dus enzyme able to modify up to three different sites as well as the first example of a tRNA-modifying enzyme that can modify bases present on the two opposite sides of an RNA-loop structure. Comparative analysis of the distribution of DusB homologs in Firmicutes revealed the existence of three DusB subgroups namely DusB1, DusB2 and DusB3. The first two subgroups were likely present in the Firmicute ancestor, and Mollicutes have retained DusB1 and lost DusB2. Altogether, our results suggest that the multisite specificity of the M. capricolum DusB enzyme could be an ancestral property.
Keywords: dihydrouridine; mollicutes; multisite-specificity; post-transcriptional modification; tRNA.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures

References
-
- Suck D, Saenger W, Zechmeister K.. Conformation of the tRNA minor constituent dihydrouridine. FEBS Lett. 1971;12:257–259. - PubMed
-
- Dyubankova N, Sochacka E, Kraszewska K, et al. Contribution of dihydrouridine in folding of the D-arm in tRNA. Org Biomol Chem. 2015;13:4960–4966. - PubMed
-
- Kim SH, Suddath FL, Quigley GJ, et al. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science. 1974;185:435–440. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
