Hyperproduction of 3-hydroxypropionate by Halomonas bluephagenesis
- PMID: 33686068
- PMCID: PMC7940609
- DOI: 10.1038/s41467-021-21632-3
Hyperproduction of 3-hydroxypropionate by Halomonas bluephagenesis
Abstract
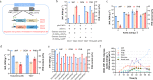
3-Hydroxypropionic acid (3HP), an important three carbon (C3) chemical, is designated as one of the top platform chemicals with an urgent need for improved industrial production. Halomonas bluephagenesis shows the potential as a chassis for competitive bioproduction of various chemicals due to its ability to grow under an open, unsterile and continuous process. Here, we report the strategy for producing 3HP and its copolymer poly(3-hydroxybutyrate-co-3-hydroxypropionate) (P3HB3HP) by the development of H. bluephagenesis. The transcriptome analysis reveals its 3HP degradation and synthesis pathways involving endogenous synthetic enzymes from 1,3-propanediol. Combing the optimized expression of aldehyde dehydrogenase (AldDHb), an engineered H. bluephagenesis strain of whose 3HP degradation pathway is deleted and that overexpresses alcohol dehydrogenases (AdhP) on its genome under a balanced redox state, is constructed with an enhanced 1.3-propanediol-dependent 3HP biosynthetic pathway to produce 154 g L-1 of 3HP with a yield and productivity of 0.93 g g-1 1,3-propanediol and 2.4 g L-1 h-1, respectively. Moreover, the strain could also accumulate 60% poly(3-hydroxybutyrate-co-32-45% 3-hydroxypropionate) in the dry cell mass, demonstrating to be a suitable chassis for hyperproduction of 3HP and P3HB3HP.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Della Pina C, Falletta E, Rossi M. A green approach to chemical building blocks. case 3-hydroxypropanoic acid. Green. Chem. 2011;13:1624–1632. doi: 10.1039/c1gc15052a. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

