Gene-expression correlates of the oscillatory signatures supporting human episodic memory encoding
- PMID: 33686299
- PMCID: PMC8016736
- DOI: 10.1038/s41593-021-00803-x
Gene-expression correlates of the oscillatory signatures supporting human episodic memory encoding
Abstract
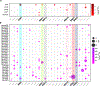
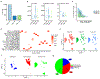
In humans, brain oscillations support critical features of memory formation. However, understanding the molecular mechanisms underlying this activity remains a major challenge. Here, we measured memory-sensitive oscillations using intracranial electroencephalography recordings from the temporal cortex of patients performing an episodic memory task. When these patients subsequently underwent resection, we employed transcriptomics on the temporal cortex to link gene expression with brain oscillations and identified genes correlated with oscillatory signatures of memory formation across six frequency bands. A co-expression analysis isolated oscillatory signature-specific modules associated with neuropsychiatric disorders and ion channel activity, with highly correlated genes exhibiting strong connectivity within these modules. Using single-nucleus transcriptomics, we further revealed that these modules are enriched for specific classes of both excitatory and inhibitory neurons, and immunohistochemistry confirmed expression of highly correlated genes. This unprecedented dataset of patient-specific brain oscillations coupled to genomics unlocks new insights into the genetic mechanisms that support memory encoding.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures





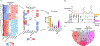




Comment in
-
Genes that give our brains their rhythms.Nat Neurosci. 2021 Apr;24(4):455-456. doi: 10.1038/s41593-021-00805-9. Nat Neurosci. 2021. PMID: 33686296 No abstract available.
References
Methods-only References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

