Single-molecule FRET imaging of GPCR dimers in living cells
- PMID: 33686301
- PMCID: PMC8232828
- DOI: 10.1038/s41592-021-01081-y
Single-molecule FRET imaging of GPCR dimers in living cells
Abstract
Class C G protein-coupled receptors (GPCRs) are known to form stable homodimers or heterodimers critical for function, but the oligomeric status of class A and B receptors, which constitute >90% of all GPCRs, remains hotly debated. Single-molecule fluorescence resonance energy transfer (smFRET) is a powerful approach with the potential to reveal valuable insights into GPCR organization but has rarely been used in living cells to study protein systems. Here, we report generally applicable methods for using smFRET to detect and track transmembrane proteins diffusing within the plasma membrane of mammalian cells. We leverage this in-cell smFRET approach to show agonist-induced structural dynamics within individual metabotropic glutamate receptor dimers. We apply these methods to representative class A, B and C receptors, finding evidence for receptor monomers, density-dependent dimers and constitutive dimers, respectively.
Conflict of interest statement
Competing interests
S.C.B. has an equity interest in Lumidyne Technologies. The other authors have no competing interests.
Figures






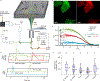
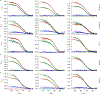

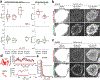





Comment in
-
Single-molecule FRET on its way to structural biology in live cells.Nat Methods. 2021 Apr;18(4):344-345. doi: 10.1038/s41592-021-01084-9. Nat Methods. 2021. PMID: 33686302 No abstract available.
References
-
- Asher WB et al. Extreme vetting of dopamine receptor oligomerization. In Receptors 33, 99–127 (Humana Press, 2017).
-
- White JH et al. Heterodimerization is required for the formation of a functional GABAB receptor. Nature 396, 679–682 (1998). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

