HIV-1 Integrase Inhibitors with Modifications That Affect Their Potencies against Drug Resistant Integrase Mutants
- PMID: 33686850
- PMCID: PMC8205226
- DOI: 10.1021/acsinfecdis.0c00819
HIV-1 Integrase Inhibitors with Modifications That Affect Their Potencies against Drug Resistant Integrase Mutants
Abstract
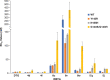
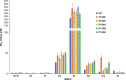
Integrase strand transfer inhibitors (INSTIs) block the integration step of the retroviral lifecycle and are first-line drugs used for the treatment of HIV-1/AIDS. INSTIs have a polycyclic core with heteroatom triads, chelate the metal ions at the active site, and have a halobenzyl group that interacts with viral DNA attached to the core by a flexible linker. The most broadly effective INSTIs inhibit both wild-type (WT) integrase (IN) and a variety of well-known mutants. However, because there are mutations that reduce the potency of all of the available INSTIs, new and better compounds are needed. Models based on recent structures of HIV-1 and red-capped mangabey SIV INs suggest modifications in the INSTI structures that could enhance interactions with the 3'-terminal adenosine of the viral DNA, which could improve performance against INSTI resistant mutants. We designed and tested a series of INSTIs having modifications to their naphthyridine scaffold. One of the new compounds retained good potency against an expanded panel of HIV-1 IN mutants that we tested. Our results suggest the possibility of designing inhibitors that combine the best features of the existing compounds, which could provide additional efficacy against known HIV-1 IN mutants.
Keywords: inhibition; integrase; mutant; potency; strand transfer; susceptibility.
Conflict of interest statement
The authors declare no competing financial interest.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

