Design of an improved universal signal peptide based on the α-factor mating secretion signal for enzyme production in yeast
- PMID: 33687500
- PMCID: PMC8038962
- DOI: 10.1007/s00018-021-03793-y
Design of an improved universal signal peptide based on the α-factor mating secretion signal for enzyme production in yeast
Abstract
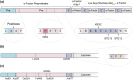
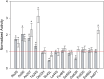
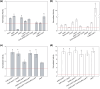
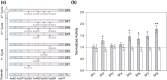
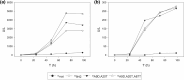
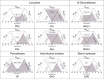
Saccharomyces cerevisiae plays an important role in the heterologous expression of an array of proteins due to its easy manipulation, low requirements and ability for protein post-translational modifications. The implementation of the preproleader secretion signal of the α-factor mating pheromone from this yeast contributes to increase the production yields by targeting the foreign protein to the extracellular environment. The use of this signal peptide combined with enzyme-directed evolution allowed us to achieve the otherwise difficult functional expression of fungal laccases in S. cerevisiae, obtaining different evolved α-factor preproleader sequences that enhance laccase secretion. However, the design of a universal signal peptide to enhance the production of heterologous proteins in S. cerevisiae is a pending challenge. We describe here the optimisation of the α-factor preproleader to improve recombinant enzyme production in S. cerevisiae through two parallel engineering strategies: a bottom-up design over the native α-factor preproleader (αnat) and a top-down design over the fittest evolved signal peptide obtained in our lab (α9H2 leader). The goal was to analyse the effect of mutations accumulated in the signal sequence throughout iterations of directed evolution, or of other reported mutations, and their possible epistatic interactions. Both approaches agreed in the positive synergism of four mutations (Aα9D, Aα20T, Lα42S, Dα83E) contained in the final optimised leader (αOPT), which notably enhanced the secretion of several fungal oxidoreductases and hydrolases. Additionally, we suggest a guideline to further drive the heterologous production of a particular enzyme based on combinatorial saturation mutagenesis of positions 86th and 87th of the αOPT leader fused to the target protein.
Keywords: Directed evolution; Enzyme heterologous expression; Saccharomyces cerevisiae; Signal peptide; Synthetic design; Α-factor preproleader.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Protein Engineering Approaches to Enhance Fungal Laccase Production in S. cerevisiae.Int J Mol Sci. 2021 Jan 25;22(3):1157. doi: 10.3390/ijms22031157. Int J Mol Sci. 2021. PMID: 33503813 Free PMC article.
-
Optimizing Pichia pastoris protein secretion: Role of N-linked glycosylation on the α-mating factor secretion signal leader.J Biotechnol. 2024 Aug 10;391:1-10. doi: 10.1016/j.jbiotec.2024.04.008. Epub 2024 Apr 16. J Biotechnol. 2024. PMID: 38636846
-
The MFα signal sequence in yeast-based protein secretion: challenges and innovations'.Appl Microbiol Biotechnol. 2025 Jun 5;109(1):138. doi: 10.1007/s00253-025-13532-z. Appl Microbiol Biotechnol. 2025. PMID: 40471355 Free PMC article. Review.
-
MFalpha signal peptide enhances the expression of cellulase eg1 gene in yeast.Appl Biochem Biotechnol. 2010 Oct;162(3):617-24. doi: 10.1007/s12010-009-8880-9. Epub 2009 Dec 19. Appl Biochem Biotechnol. 2010. PMID: 20024677
-
Secretion of heterologous proteins directed by the yeast alpha-factor leader.Biotechnology. 1989;13:269-80. Biotechnology. 1989. PMID: 2679928 Review. No abstract available.
Cited by
-
Fungal Laccases: Fundamentals, Engineering and Classification Update.Biomolecules. 2023 Nov 28;13(12):1716. doi: 10.3390/biom13121716. Biomolecules. 2023. PMID: 38136587 Free PMC article. Review.
-
Comprehensive Analysis of Signal Peptides in Saccharomyces cerevisiae Reveals Features for Efficient Secretion.Adv Sci (Weinh). 2023 Jan;10(2):e2203433. doi: 10.1002/advs.202203433. Epub 2022 Dec 7. Adv Sci (Weinh). 2023. PMID: 36478443 Free PMC article.
-
Saccharomyces cerevisiae cell surface display technology: Strategies for improvement and applications.Front Bioeng Biotechnol. 2022 Dec 7;10:1056804. doi: 10.3389/fbioe.2022.1056804. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 36568309 Free PMC article. Review.
-
A Yeast Modular Cloning (MoClo) Toolkit Expansion for Optimization of Heterologous Protein Secretion and Surface Display in Saccharomyces cerevisiae.ACS Synth Biol. 2024 Apr 19;13(4):1246-1258. doi: 10.1021/acssynbio.3c00743. Epub 2024 Mar 14. ACS Synth Biol. 2024. PMID: 38483353 Free PMC article.
-
Engineering strategies for enhanced heterologous protein production by Saccharomyces cerevisiae.Microb Cell Fact. 2024 Jan 22;23(1):32. doi: 10.1186/s12934-024-02299-z. Microb Cell Fact. 2024. PMID: 38247006 Free PMC article. Review.
References
-
- Huang M, Bao J, Nielsen J. Biopharmaceutical protein production by Saccharomyces cerevisiae : current state and future prospects. Pharm Bioprocess. 2014;2:167–182. doi: 10.4155/pbp.14.8. - DOI
-
- Rai M, Padh H. Expression systems for production of heterologous proteins. Curr Sci. 2001;80:1121–1128.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

