B Lymphocytes, but Not Dendritic Cells, Efficiently HIV-1 Trans Infect Naive CD4+ T Cells: Implications for the Viral Reservoir
- PMID: 33688006
- PMCID: PMC8092276
- DOI: 10.1128/mBio.02998-20
B Lymphocytes, but Not Dendritic Cells, Efficiently HIV-1 Trans Infect Naive CD4+ T Cells: Implications for the Viral Reservoir
Abstract
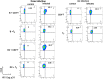
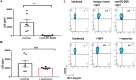
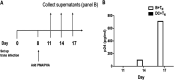
Insight into the establishment and maintenance of HIV-1 infection in resting CD4+ T cell subsets is critical for the development of therapeutics targeting the HIV-1 reservoir. Although the frequency of HIV-1 infection, as quantified by the frequency of HIV-1 DNA, is lower in CD4+ naive T cells (TN) than in the memory T cell subsets, recent studies have shown that TN harbor a large pool of replication-competent virus. Interestingly, however, TN are highly resistant to direct (cis) HIV-1 infection in vitro, in particular to R5-tropic HIV-1, as TN do not express CCR5. In this study, we investigated whether TN could be efficiently HIV-1 trans infected by professional antigen-presenting B lymphocytes and myeloid dendritic cells (DC) in the absence of global T cell activation. We found that B cells, but not DC, have a unique ability to efficiently trans infect TNin vitro In contrast, both B cells and DC mediated HIV-1 trans infection of memory and activated CD4+ T cells. Moreover, we found that TN isolated from HIV-1-infected nonprogressors (NP) harbor significantly disproportionately lower levels of HIV-1 DNA than TN isolated from progressors. This is consistent with our previous finding that antigen-presenting cells (APC) derived from NP do not efficiently trans infect CD4+ T cells due to alterations in APC cholesterol metabolism and cell membrane lipid raft organization. These findings support that B cell-mediated trans infection of TN with HIV-1 has a more profound role than previously considered in establishing the viral reservoir and control of HIV-1 disease progression.IMPORTANCE The latent human immunodeficiency virus type 1 (HIV-1) reservoir in persons on antiretroviral therapy (ART) represents a major barrier to a cure. Although most studies have focused on the HIV-1 reservoir in the memory T cell subset, replication-competent HIV-1 has been isolated from TN, and CCR5-tropic HIV-1 has been recovered from CCR5neg TN from ART-suppressed HIV-1-infected individuals. In this study, we showed that CCR5neg TN are efficiently trans infected with R5-tropic HIV-1 by B lymphocytes, but not by myeloid dendritic cells. Furthermore, we found that TN isolated from NP harbor no or significantly fewer copies of HIV-1 DNA than those from ART-suppressed progressors. These findings support that B cell-mediated trans infection of TN with HIV-1 has a more profound role than previously considered in establishing the viral reservoir and control of HIV-1 disease progression. Understanding the establishment and maintenance of the HIV-1 latent reservoir is fundamental for the design of effective treatments for viral eradication.
Keywords: B lymphocytes; HIV-1; dendritic cells; naive CD4+ T cells; trans infection.
Copyright © 2021 Gerberick et al.
Figures
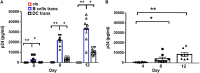


Similar articles
-
Cholesterol Metabolism in Antigen-Presenting Cells and HIV-1 Trans-Infection of CD4+ T Cells.Viruses. 2023 Nov 29;15(12):2347. doi: 10.3390/v15122347. Viruses. 2023. PMID: 38140588 Free PMC article. Review.
-
Alterations in cholesterol metabolism restrict HIV-1 trans infection in nonprogressors.mBio. 2014 Apr 29;5(3):e01031-13. doi: 10.1128/mBio.01031-13. mBio. 2014. PMID: 24781743 Free PMC article.
-
A CCR5+ memory subset within HIV-1-infected primary resting CD4+ T cells is permissive for replication-competent, latently infected viruses in vitro.BMC Res Notes. 2019 Apr 29;12(1):242. doi: 10.1186/s13104-019-4281-5. BMC Res Notes. 2019. PMID: 31036079 Free PMC article.
-
Inefficient HIV-1 trans Infection of CD4+ T Cells by Macrophages from HIV-1 Nonprogressors Is Associated with Altered Membrane Cholesterol and DC-SIGN.J Virol. 2018 Jun 13;92(13):e00092-18. doi: 10.1128/JVI.00092-18. Print 2018 Jul 1. J Virol. 2018. PMID: 29643243 Free PMC article.
-
Reservoirs for HIV-1: mechanisms for viral persistence in the presence of antiviral immune responses and antiretroviral therapy.Annu Rev Immunol. 2000;18:665-708. doi: 10.1146/annurev.immunol.18.1.665. Annu Rev Immunol. 2000. PMID: 10837072 Review.
Cited by
-
People with HIV have higher percentages of circulating CCR5+ CD8+ T cells and lower percentages of CCR5+ regulatory T cells.Sci Rep. 2022 Jul 6;12(1):11425. doi: 10.1038/s41598-022-15646-0. Sci Rep. 2022. PMID: 35794176 Free PMC article.
-
Naive infection predicts reservoir diversity and is a formidable hurdle to HIV eradication.JCI Insight. 2021 Aug 23;6(16):e150794. doi: 10.1172/jci.insight.150794. JCI Insight. 2021. PMID: 34228640 Free PMC article.
-
Cholesterol Metabolism in Antigen-Presenting Cells and HIV-1 Trans-Infection of CD4+ T Cells.Viruses. 2023 Nov 29;15(12):2347. doi: 10.3390/v15122347. Viruses. 2023. PMID: 38140588 Free PMC article. Review.
-
Antigen Presenting Cell-Mediated HIV-1 Trans Infection in the Establishment and Maintenance of the Viral Reservoir.Med Res Arch. 2023 Jul;11(7.1):10.18103/mra.v11i7.1.4064. doi: 10.18103/mra.v11i7.1.4064. Epub 2023 Jul 6. Med Res Arch. 2023. PMID: 39634038 Free PMC article.
References
-
- Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, Quinn TC, Chaisson RE, Rosenberg E, Walker B, Gange S, Gallant J, Siliciano RF. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med 5:512–517. doi:10.1038/8394. - DOI - PubMed
-
- Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295–1300. doi:10.1126/science.278.5341.1295. - DOI - PubMed
-
- Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel MR, Ghattas G, Brenchley JM, Schacker TW, Hill BJ, Douek DC, Routy JP, Haddad EK, Sekaly RP. 2009. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med 15:893–900. doi:10.1038/nm.1972. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

