SARS-CoV-2 within-host diversity and transmission
- PMID: 33688063
- PMCID: PMC8128293
- DOI: 10.1126/science.abg0821
SARS-CoV-2 within-host diversity and transmission
Abstract
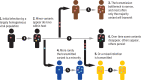
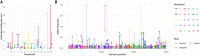
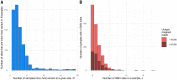
Extensive global sampling and sequencing of the pandemic virus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have enabled researchers to monitor its spread and to identify concerning new variants. Two important determinants of variant spread are how frequently they arise within individuals and how likely they are to be transmitted. To characterize within-host diversity and transmission, we deep-sequenced 1313 clinical samples from the United Kingdom. SARS-CoV-2 infections are characterized by low levels of within-host diversity when viral loads are high and by a narrow bottleneck at transmission. Most variants are either lost or occasionally fixed at the point of transmission, with minimal persistence of shared diversity, patterns that are readily observable on the phylogenetic tree. Our results suggest that transmission-enhancing and/or immune-escape SARS-CoV-2 variants are likely to arise infrequently but could spread rapidly if successfully transmitted.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures



References
-
- Lu J., du Plessis L., Liu Z., Hill V., Kang M., Lin H., Sun J., Franois S., Kraemer M. U. G., Faria N. R., McCrone J. T., Peng J., Xiong Q., Yuan R., Zeng L., Zhou P., Liang C., Yi L., Liu J., Xiao J., Hu J., Liu T., Ma W., Li W., Su J., Zheng H., Peng B., Fang S., Su W., Li K., Sun R., Bai R., Tang X., Liang M., Quick J., Song T., Rambaut A., Loman N., Raghwani J., Pybus O. G., Ke C., Genomic epidemiology of SARS-CoV-2 in Guangdong Province, China. Cell 181, 997–1003.e9. (2020). 10.1016/j.cell.2020.04.023 - DOI - PMC - PubMed
-
- Korber B., Fischer W. M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E. E., Bhattacharya T., Foley B., Hastie K. M., Parker M. D., Partridge D. G., Evans C. M., Freeman T. M., de Silva T. I., McDanal C., Perez L. G., Tang H., Moon-Walker A., Whelan S. P., LaBranche C. C., Saphire E. O., Montefiori D. C., Angyal A., Brown R. L., Carrilero L., Green L. R., Groves D. C., Johnson K. J., Keeley A. J., Lindsey B. B., Parsons P. J., Raza M., Rowland-Jones S., Smith N., Tucker R. M., Wang D., Wyles M. D.; Sheffield COVID-19 Genomics Group , Tracking changes in SARS-CoV-2 spike: Evidence that D614G increases infectivity of the COVID-19 virus. Cell 182, 812–827.e19. (2020). 10.1016/j.cell.2020.06.043 - DOI - PMC - PubMed
-
- Volz E., Hill V., McCrone J. T., Price A., Jorgensen D., OToole ., Southgate J., Johnson R., Jackson B., Nascimento F. F., Rey S. M., Nicholls S. M., Colquhoun R. M., da Silva Filipe A., Shepherd J., Pascall D. J., Shah R., Jesudason N., Li K., Jarrett R., Pacchiarini N., Bull M., Geidelberg L., Siveroni I., Goodfellow I., Loman N. J., Pybus O. G., Robertson D. L., Thomson E. C., Rambaut A., Connor T. R., Koshy C., Wise E., Cortes N., Lynch J., Kidd S., Mori M., Fairley D. J., Curran T., McKenna J. P., Adams H., Fraser C., Golubchik T., Bonsall D., Moore C., Caddy S. L., Khokhar F. A., Wantoch M., Reynolds N., Warne B., Maksimovic J., Spellman K., McCluggage K., John M., Beer R., Afifi S., Morgan S., Marchbank A., Price A., Kitchen C., Gulliver H., Merrick I., Southgate J., Guest M., Munn R., Workman T., Connor T. R., Fuller W., Bresner C., Snell L. B., Charalampous T., Nebbia G., Batra R., Edgeworth J., Robson S. C., Beckett A., Loveson K. F., Aanensen D. M., Underwood A. P., Yeats C. A., Abudahab K., Taylor B. E. W., Menegazzo M., Clark G., Smith W., Khakh M., Fleming V. M., Lister M. M., Howson-Wells H. C., Berry L., Boswell T., Joseph A., Willingham I., Bird P., Helmer T., Fallon K., Holmes C., Tang J., Raviprakash V., Campbell S., Sheriff N., Loose M. W., Holmes N., Moore C., Carlile M., Wright V., Sang F., Debebe J., Coll F., Signell A. W., Betancor G., Wilson H. D., Feltwell T., Houldcroft C. J., Eldirdiri S., Kenyon A., Davis T., Pybus O., du Plessis L., Zarebski A., Raghwani J., Kraemer M., Francois S., Attwood S., Vasylyeva T., Torok M. E., Hamilton W. L., Goodfellow I. G., Hall G., Jahun A. S., Chaudhry Y., Hosmillo M., Pinckert M. L., Georgana I., Yakovleva A., Meredith L. W., Moses S., Lowe H., Ryan F., Fisher C. L., Awan A. R., Boyes J., Breuer J., Harris K. A., Brown J. R., Shah D., Atkinson L., Lee J. C. D., Alcolea-Medina A., Moore N., Cortes N., Williams R., Chapman M. R., Levett L. J., Heaney J., Smith D. L., Bashton M., Young G. R., Allan J., Loh J., Randell P. A., Cox A., Madona P., Holmes A., Bolt F., Price J., Mookerjee S., Rowan A., Taylor G. P., Ragonnet-Cronin M., Nascimento F. F., Jorgensen D., Siveroni I., Johnson R., Boyd O., Geidelberg L., Volz E. M., Brunker K., Smollett K. L., Loman N. J., Quick J., McMurray C., Stockton J., Nicholls S., Rowe W., Poplawski R., Martinez-Nunez R. T., Mason J., Robinson T. I., OToole E., Watts J., Breen C., Cowell A., Ludden C., Sluga G., Machin N. W., Ahmad S. S. Y., George R. P., Halstead F., Sivaprakasam V., Thomson E. C., Shepherd J. G., Asamaphan P., Niebel M. O., Li K. K., Shah R. N., Jesudason N. G., Parr Y. A., Tong L., Broos A., Mair D., Nichols J., Carmichael S. N., Nomikou K., Aranday-Cortes E., Johnson N., Starinskij I., da Silva Filipe A., Robertson D. L., Orton R. J., Hughes J., Vattipally S., Singer J. B., Hale A. D., Macfarlane-Smith L. R., Harper K. L., Taha Y., Payne B. A. I., Burton-Fanning S., Waugh S., Collins J., Eltringham G., Templeton K. E., McHugh M. P., Dewar R., Wastenge E., Dervisevic S., Stanley R., Prakash R., Stuart C., Elumogo N., Sethi D. K., Meader E. J., Coupland L. J., Potter W., Graham C., Barton E., Padgett D., Scott G., Swindells E., Greenaway J., Nelson A., Yew W. C., Resende Silva P. C., Andersson M., Shaw R., Peto T., Justice A., Eyre D., Crooke D., Hoosdally S., Sloan T. J., Duckworth N., Walsh S., Chauhan A. J., Glaysher S., Bicknell K., Wyllie S., Butcher E., Elliott S., Lloyd A., Impey R., Levene N., Monaghan L., Bradley D. T., Allara E., Pearson C., Muir P., Vipond I. B., Hopes R., Pymont H. M., Hutchings S., Curran M. D., Parmar S., Lackenby A., Mbisa T., Platt S., Miah S., Bibby D., Manso C., Hubb J., Chand M., Dabrera G., Ramsay M., Bradshaw D., Thornton A., Myers R., Schaefer U., Groves N., Gallagher E., Lee D., Williams D., Ellaby N., Harrison I., Hartman H., Manesis N., Patel V., Bishop C., Chalker V., Osman H., Bosworth A., Robinson E., Holden M. T. G., Shaaban S., Birchley A., Adams A., Davies A., Gaskin A., Plimmer A., Gatica-Wilcox B., McKerr C., Moore C., Williams C., Heyburn D., De Lacy E., Hilvers E., Downing F., Shankar G., Jones H., Asad H., Coombes J., Watkins J., Evans J. M., Fina L., Gifford L., Gilbert L., Graham L., Perry M., Morgan M., Bull M., Cronin M., Pacchiarini N., Craine N., Jones R., Howe R., Corden S., Rey S., Kumziene-Summerhayes S., Taylor S., Cottrell S., Jones S., Edwards S., OGrady J., Page A. J., Wain J., Webber M. A., Mather A. E., Baker D. J., Rudder S., Yasir M., Thomson N. M., Aydin A., Tedim A. P., Kay G. L., Trotter A. J., Gilroy R. A. J., Alikhan N.-F., de Oliveira Martins L., Le-Viet T., Meadows L., Kolyva A., Diaz M., Bell A., Gutierrez A. V., Charles I. G., Adriaenssens E. M., Kingsley R. A., Casey A., Simpson D. A., Molnar Z., Thompson T., Acheson E., Masoli J. A. H., Knight B. A., Hattersley A., Ellard S., Auckland C., Mahungu T. W., Irish-Tavares D., Haque T., Bourgeois Y., Scarlett G. P., Partridge D. G., Raza M., Evans C., Johnson K., Liggett S., Baker P., Essex S., Lyons R. A., Caller L. G., Castellano S., Williams R. J., Kristiansen M., Roy S., Williams C. A., Dyal P. L., Tutill H. J., Panchbhaya Y. N., Forrest L. M., Niola P., Findlay J., Brooks T. T., Gavriil A., Mestek-Boukhibar L., Weeks S., Pandey S., Berry L., Jones K., Richter A., Beggs A., Smith C. P., Bucca G., Hesketh A. R., Harrison E. M., Peacock S. J., Palmer S., Churcher C. M., Bellis K. L., Girgis S. T., Naydenova P., Blane B., Sridhar S., Ruis C., Forrest S., Cormie C., Gill H. K., Dias J., Higginson E. E., Maes M., Young J., Kermack L. M., Hadjirin N. F., Aggarwal D., Griffith L., Swingler T., Davidson R. K., Rambaut A., Williams T., Balcazar C. E., Gallagher M. D., OToole ., Rooke S., Jackson B., Colquhoun R., Ashworth J., Hill V., McCrone J. T., Scher E., Yu X., Williamson K. A., Stanton T. D., Michell S. L., Bewshea C. M., Temperton B., Michelsen M. L., Warwick-Dugdale J., Manley R., Farbos A., Harrison J. W., Sambles C. M., Studholme D. J., Jeffries A. R., Darby A. C., Hiscox J. A., Paterson S., Iturriza-Gomara M., Jackson K. A., Lucaci A. O., Vamos E. E., Hughes M., Rainbow L., Eccles R., Nelson C., Whitehead M., Turtle L., Haldenby S. T., Gregory R., Gemmell M., Kwiatkowski D., de Silva T. I., Smith N., Angyal A., Lindsey B. B., Groves D. C., Green L. R., Wang D., Freeman T. M., Parker M. D., Keeley A. J., Parsons P. J., Tucker R. M., Brown R., Wyles M., Constantinidou C., Unnikrishnan M., Ott S., Cheng J. K. J., Bridgewater H. E., Frost L. R., Taylor-Joyce G., Stark R., Baxter L., Alam M. T., Brown P. E., McClure P. C., Chappell J. G., Tsoleridis T., Ball J., Gramatopoulos D., Buck D., Todd J. A., Green A., Trebes A., MacIntyre-Cockett G., de Cesare M., Langford C., Alderton A., Amato R., Goncalves S., Jackson D. K., Johnston I., Sillitoe J., Palmer S., Lawniczak M., Berriman M., Danesh J., Livett R., Shirley L., Farr B., Quail M., Thurston S., Park N., Betteridge E., Weldon D., Goodwin S., Nelson R., Beaver C., Letchford L., Jackson D. A., Foulser L., McMinn L., Prestwood L., Kay S., Kane L., Dorman M. J., Martincorena I., Puethe C., Keatley J.-P., Tonkin-Hill G., Smith C., Jamrozy D., Beale M. A., Patel M., Ariani C., Spencer-Chapman M., Drury E., Lo S., Rajatileka S., Scott C., James K., Buddenborg S. K., Berger D. J., Patel G., Garcia-Casado M. V., Dibling T., McGuigan S., Rogers H. A., Hunter A. D., Souster E., Neaverson A. S.; COG-UK Consortium , Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell 184, 64–75.e11. (2021). 10.1016/j.cell.2020.11.020 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

