A single-plasmid approach for genome editing coupled with long-term lineage analysis in chick embryos
- PMID: 33688075
- PMCID: PMC8077534
- DOI: 10.1242/dev.193565
A single-plasmid approach for genome editing coupled with long-term lineage analysis in chick embryos
Erratum in
-
Correction: A single-plasmid approach for genome editing coupled with long-term lineage analysis in chick embryos.Development. 2022 Jul 1;149(13):dev201078. doi: 10.1242/dev.201078. Epub 2022 Jul 11. Development. 2022. PMID: 35819067 Free PMC article. No abstract available.
Abstract
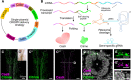
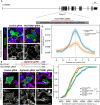
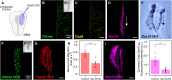
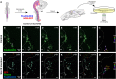
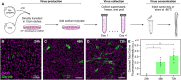
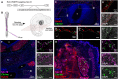
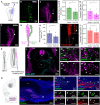
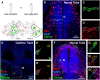
An important strategy for establishing mechanisms of gene function during development is through mutation of individual genes and analysis of subsequent effects on cell behavior. Here, we present a single-plasmid approach for genome editing in chick embryos to study experimentally perturbed cells in an otherwise normal embryonic environment. To achieve this, we have engineered a plasmid that encodes Cas9 protein, gene-specific guide RNA (gRNA), and a fluorescent marker within the same construct. Using transfection- and electroporation-based approaches, we show that this construct can be used to perturb gene function in early embryos as well as human cell lines. Importantly, insertion of this cistronic construct into replication-incompetent avian retroviruses allowed us to couple gene knockouts with long-term lineage analysis. We demonstrate the application of our newly engineered constructs and viruses by perturbing β-catenin in vitro and Sox10, Pax6 and Pax7 in the neural crest, retina, and neural tube and segmental plate in vivo, respectively. Together, this approach enables genes of interest to be knocked out in identifiable cells in living embryos and can be broadly applied to numerous genes in different embryonic tissues.
Keywords: CRISPR-Cas9; Chick embryology; Clonal analysis; Genome editing; Migration; Neural crest; Retroviruses.
© 2021. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures

Comment in
-
A new construct for CRISPR delivery in chicken.Lab Anim (NY). 2021 May;50(5):120. doi: 10.1038/s41684-021-00765-4. Lab Anim (NY). 2021. PMID: 33911254 No abstract available.
References
-
- Andreason, G. L. and Evans, G. A. (1988). Introduction and expression of DNA molecules in eukaryotic cells by electroporation. BioTechniques 6, 650-660. - PubMed
-
- Bernsen, J. (1986). Dynamic thresholding of grey-level images. In Proceedings of the Eighth International Conference on Pattern Recognition, pp. 1251-1255. IEEE Computer Society Press.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

