A Microfluidic Method for the Selection of Undifferentiated Human Embryonic Stem Cells and in Situ Analysis
- PMID: 33688311
- PMCID: PMC7939131
- DOI: 10.1007/s10404-014-1485-9
A Microfluidic Method for the Selection of Undifferentiated Human Embryonic Stem Cells and in Situ Analysis
Abstract
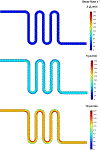
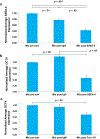
Conventional cell-sorting methods such as fluorescence-activated cell sorting (FACS) or magnetic-activated cell sorting (MACS) can suffer from certain shortcomings such as lengthy sample preparation time, cell modification through antibody labeling, and cell damage due to exposure to high shear forces or to attachment of superparamagnetic Microbeads. In light of these drawbacks, we have recently developed a label-free, microfluidic platform that can not only select cells with minimal sample preparation but also enable analysis of cells in situ. We demonstrate the utility of our platform by successfully isolating undifferentiated human embryonic stem cells (hESCs) from a heterogeneous population based on the undifferentiated stem-cell marker SSEA-4. Importantly, we show that, in contrast to MACS or FACS, cells isolated by our method have very high viability (~90%). Overall, our platform technology could likely be applied to other cell types beyond hESCs and to a variety of heterogeneous cell populations in order to select and analyze cells of interest.
Keywords: Human embryonic stem cells (hESCs); cell sorting; in situ analysis; label-free.
Figures




Similar articles
-
Separation of SSEA-4 and TRA-1-60 labelled undifferentiated human embryonic stem cells from a heterogeneous cell population using magnetic-activated cell sorting (MACS) and fluorescence-activated cell sorting (FACS).Stem Cell Rev Rep. 2009 Mar;5(1):72-80. doi: 10.1007/s12015-009-9054-4. Epub 2009 Jan 31. Stem Cell Rev Rep. 2009. PMID: 19184635
-
[Comparison of sorting of fluorescently and magnetically labelled dental pulp stem cells].Fogorv Sz. 2016 Mar;109(1):29-33. Fogorv Sz. 2016. PMID: 27188159 Hungarian.
-
Evaluation of plasma cell sorting methods in multiple myeloma patients: flow cytometry versus magnetic beads.Cancer Cell Int. 2025 Jan 17;25(1):16. doi: 10.1186/s12935-025-03647-8. Cancer Cell Int. 2025. PMID: 39825397 Free PMC article.
-
Label-free cell sorting strategies via biophysical and biochemical gradients.J Orthop Translat. 2019 Feb 26;17:55-63. doi: 10.1016/j.jot.2019.01.005. eCollection 2019 Apr. J Orthop Translat. 2019. PMID: 31194093 Free PMC article. Review.
-
Recent advances in microfluidic cell sorting techniques based on both physical and biochemical principles.Electrophoresis. 2019 Mar;40(6):930-954. doi: 10.1002/elps.201800361. Epub 2018 Oct 23. Electrophoresis. 2019. PMID: 30311661 Review.
Cited by
-
Micro/nanoengineered technologies for human pluripotent stem cells maintenance and differentiation.Nano Today. 2021 Dec;41:101310. doi: 10.1016/j.nantod.2021.101310. Epub 2021 Oct 6. Nano Today. 2021. PMID: 34745321 Free PMC article.
References
-
- Abcam. (n.d.). Indirect flow cytometry (FACS) protocol. Retrieved from http://www.abcam.com/index.html?pageconfig=resource&rid=11381
-
- Akerstroms B, & Bjork L (1989). Protein L : An Immunoglobulin Light Chain-binding Bacterial Protein, 264(33), 19740–19746. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
