The Identification and Validation of a Robust Immune-Associated Gene Signature in Cutaneous Melanoma
- PMID: 33688507
- PMCID: PMC7911606
- DOI: 10.1155/2021/6686284
The Identification and Validation of a Robust Immune-Associated Gene Signature in Cutaneous Melanoma
Abstract
Background: Cutaneous melanoma is defined as one of the most aggressive skin tumors in the world. An increasing body of evidence suggested an indispensable association between immune-associated gene (IAG) signature and melanoma. This article is aimed at formulating an IAG signature to estimate prognosis of melanoma.
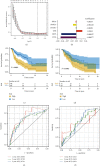
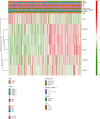
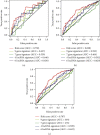
Methods: 434 melanoma patients were extracted from The Cancer Genome Atlas (TCGA) database, and 1811 IAGs were downloaded from the ImmPort database in our retrospective study. The Cox regression analysis and LASSO regression analysis were utilized to establish a prognostic IAG signature. The Kaplan-Meier (KM) survival analysis was performed, and the time-dependent receiver operating characteristic curve (ROC) analysis was further applied to assess the predictive value. Besides, the propensity score algorithm was utilized to balance the confounding clinical factors between the high- and low-risk groups.
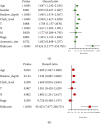
Results: A total of six prognostic IAGs comprising of INHA, NDRG1, IFITM1, LHB, GBP2, and CCL8 were eventually filtered out. According to the KM survival analysis, the results displayed a shorter overall survival (OS) in the high-risk group compared to the low-risk group. In the multivariate Cox model, the gene signature was testified as a remarkable prognostic factor (HR = 45.423, P < 0.001). Additionally, the ROC curve analyses were performed which demonstrated our IAG signature was superior to four known biomarkers mentioned in the study. Moreover, the IAG signature was significantly related to immunotherapy-related biomarkers.
Conclusion: Our study demonstrated that the six IAG signature played a critical role in the prognosis and immunotherapy of melanoma, which might help clinicians predict patients' survival and provide individualized treatment.
Copyright © 2021 Le-Bin Song et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures
Similar articles
-
Predicting the clinical outcome of melanoma using an immune-related gene pairs signature.PLoS One. 2020 Oct 8;15(10):e0240331. doi: 10.1371/journal.pone.0240331. eCollection 2020. PLoS One. 2020. PMID: 33031392 Free PMC article.
-
Melanoma long non-coding RNA signature predicts prognostic survival and directs clinical risk-specific treatments.J Dermatol Sci. 2017 Mar;85(3):226-234. doi: 10.1016/j.jdermsci.2016.12.006. Epub 2016 Dec 5. J Dermatol Sci. 2017. PMID: 27955882
-
Identification of immune-associated gene signature and immune cell infiltration related to overall survival in progressive multiple sclerosis.Mult Scler Relat Disord. 2021 Oct;55:103188. doi: 10.1016/j.msard.2021.103188. Epub 2021 Aug 2. Mult Scler Relat Disord. 2021. PMID: 34371273
-
A Novel Pancreatic Cancer Hypoxia Status Related Gene Signature for Prognosis and Therapeutic Responses.Mol Biotechnol. 2024 Jul;66(7):1684-1703. doi: 10.1007/s12033-023-00807-x. Epub 2023 Jul 5. Mol Biotechnol. 2024. PMID: 37405638 Review.
-
Emerging prognostic biomarkers in advanced cutaneous melanoma: a literature update.Expert Rev Mol Diagn. 2024 Jan-Feb;24(1-2):49-66. doi: 10.1080/14737159.2024.2314574. Epub 2024 Feb 9. Expert Rev Mol Diagn. 2024. PMID: 38334382 Review.
Cited by
-
Machine learning-based construction of immunogenic cell death-related score for improving prognosis and response to immunotherapy in melanoma.Aging (Albany NY). 2023 Apr 6;15(7):2667-2688. doi: 10.18632/aging.204636. Epub 2023 Apr 6. Aging (Albany NY). 2023. PMID: 37036471 Free PMC article.
-
Construction and Identification of an NLR-Associated Prognostic Signature Revealing the Heterogeneous Immune Response in Skin Cutaneous Melanoma.Clin Cosmet Investig Dermatol. 2023 Jun 26;16:1623-1639. doi: 10.2147/CCID.S410723. eCollection 2023. Clin Cosmet Investig Dermatol. 2023. PMID: 37396711 Free PMC article.
-
Crosstalk of four kinds of cell deaths defines subtypes of cutaneous melanoma for precise immunotherapy and chemotherapy.Front Immunol. 2022 Nov 30;13:998454. doi: 10.3389/fimmu.2022.998454. eCollection 2022. Front Immunol. 2022. PMID: 36532053 Free PMC article.
-
What genes are differentially expressed in individuals with schizophrenia? A systematic review.Mol Psychiatry. 2022 Mar;27(3):1373-1383. doi: 10.1038/s41380-021-01420-7. Epub 2022 Jan 28. Mol Psychiatry. 2022. PMID: 35091668 Free PMC article.
-
Explore the impact of hypoxia-related genes (HRGs) in Cutaneous melanoma.BMC Med Genomics. 2023 Jul 8;16(1):160. doi: 10.1186/s12920-023-01587-8. BMC Med Genomics. 2023. PMID: 37422626 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

