This is a preprint.
Systematic analysis of SARS-CoV-2 infection of an ACE2-negative human airway cell
- PMID: 33688646
- PMCID: PMC7941617
- DOI: 10.1101/2021.03.01.433431
Systematic analysis of SARS-CoV-2 infection of an ACE2-negative human airway cell
Update in
-
Systematic analysis of SARS-CoV-2 infection of an ACE2-negative human airway cell.Cell Rep. 2021 Jul 13;36(2):109364. doi: 10.1016/j.celrep.2021.109364. Epub 2021 Jun 23. Cell Rep. 2021. PMID: 34214467 Free PMC article.
Abstract
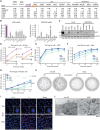
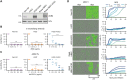
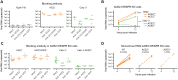
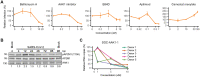
Established in vitro models for SARS-CoV-2 infection are limited and include cell lines of non-human origin and those engineered to overexpress ACE2, the cognate host cell receptor. We identified human H522 lung adenocarcinoma cells as naturally permissive to SARS-CoV-2 infection despite complete absence of ACE2. Infection of H522 cells required the SARS-CoV-2 spike protein, though in contrast to ACE2-dependent models, spike alone was not sufficient for H522 infection. Temporally resolved transcriptomic and proteomic profiling revealed alterations in cell cycle and the antiviral host cell response, including MDA5-dependent activation of type-I interferon signaling. Focused chemical screens point to important roles for clathrin-mediated endocytosis and endosomal cathepsins in SARS-CoV-2 infection of H522 cells. These findings imply the utilization of an alternative SARS-CoV-2 host cell receptor which may impact tropism of SARS-CoV-2 and consequently human disease pathogenesis.
Conflict of interest statement
DECLARATION OF INTERESTS
S.P.J.W., P.W.R. and Washington University have filed a patent application for uses of VSV-SARS-CoV-2. S.P.J.W has received unrelated funding support in sponsored research agreements with Vir Biotechnology, Abbvie and SAB therapeutics.
Figures


References
-
- Agajanian M.J., Walker M.P., Axtman A.D., Ruela-de-Sousa R.R., Serafin D.S., Rabinowitz A.D., Graham D.M., Ryan M.B., Tamir T., Nakamichi Y., et al. (2019). WNT Activates the AAK1 Kinase to Promote Clathrin-Mediated Endocytosis of LRP6 and Establish a Negative Feedback Loop. Cell Rep 26, 79–93 e78. - PMC - PubMed
-
- Aguiar J.A., Tremblay B.J., Mansfield M.J., Woody O., Lobb B., Banerjee A., Chandiramohan A., Tiessen N., Cao Q., Dvorkin-Gheva A., et al. (2020). Gene expression and in situ protein profiling of candidate SARS-CoV-2 receptors in human airway epithelial cells and lung tissue. Eur Respir J 56. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
